Protein S-thiolation by Glutathionylspermidine (Gsp): the role of Escherichia coli Gsp synthetASE/amidase in redox regulation
- PMID: 20530482
- PMCID: PMC2919097
- DOI: 10.1074/jbc.M110.133363
Protein S-thiolation by Glutathionylspermidine (Gsp): the role of Escherichia coli Gsp synthetASE/amidase in redox regulation
Abstract
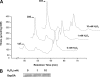
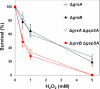
Certain bacteria synthesize glutathionylspermidine (Gsp), from GSH and spermidine. Escherichia coli Gsp synthetase/amidase (GspSA) catalyzes both the synthesis and hydrolysis of Gsp. Prior to the work reported herein, the physiological role(s) of Gsp or how the two opposing GspSA activities are regulated had not been elucidated. We report that Gsp-modified proteins from E. coli contain mixed disulfides of Gsp and protein thiols, representing a new type of post-translational modification formerly undocumented. The level of these proteins is increased by oxidative stress. We attribute the accumulation of such proteins to the selective inactivation of GspSA amidase activity. X-ray crystallography and a chemical modification study indicated that the catalytic cysteine thiol of the GspSA amidase domain is transiently inactivated by H(2)O(2) oxidation to sulfenic acid, which is stabilized by a very short hydrogen bond with a water molecule. We propose a set of reactions that explains how the levels of Gsp and Gsp S-thiolated proteins are modulated in response to oxidative stress. The hypersensitivities of GspSA and GspSA/glutaredoxin null mutants to H(2)O(2) support the idea that GspSA and glutaredoxin act synergistically to regulate the redox environment of E. coli.
Figures

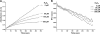




References
-
- Winterbourn C. C., Hampton M. B. (2008) Free Radic. Biol. Med. 45, 549–561 - PubMed
-
- Ritz D., Beckwith J. (2001) Annu. Rev. Microbiol. 55, 21–48 - PubMed
-
- Dalle-Donne I., Rossi R., Giustarini D., Colombo R., Milzani A. (2007) Free Radic. Biol. Med. 43, 883–898 - PubMed
-
- Dalle-Donne I., Rossi R., Colombo G., Giustarini D., Milzani A. (2009) Trends Biochem. Sci. 34, 85–96 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

