Synonymous codon usage influences the local protein structure observed
- PMID: 20530529
- PMCID: PMC2965230
- DOI: 10.1093/nar/gkq495
Synonymous codon usage influences the local protein structure observed
Abstract
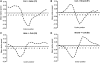
Translation of mRNA into protein is a unidirectional information flow process. Analysing the input (mRNA) and output (protein) of translation, we find that local protein structure information is encoded in the mRNA nucleotide sequence. The Coding Sequence and Structure (CSandS) database developed in this work provides a detailed mapping between over 4000 solved protein structures and their mRNA. CSandS facilitates a comprehensive analysis of codon usage over many organisms. In assigning translation speed, we find that relative codon usage is less informative than tRNA concentration. For all speed measures, no evidence was found that domain boundaries are enriched with slow codons. In fact, genes seemingly avoid slow codons around structurally defined domain boundaries. Translation speed, however, does decrease at the transition into secondary structure. Codons are identified that have structural preferences significantly different from the amino acid they encode. However, each organism has its own set of 'significant codons'. Our results support the premise that codons encode more information than merely amino acids and give insight into the role of translation in protein folding.
Figures

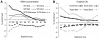
References
-
- Jones DT. Progress in protein structure prediction. Curr. Opin. Struct. Biol. 1997;7:377–387. - PubMed
-
- Koehl P, Levitt M. A brighter future for protein structure prediction. Nat. Struct. Mol. Biol. 1999;6:108–111. - PubMed
-
- Moult J. Predicting protein three-dimensional structure. Curr. Opin. Biotechnol. 1999;10:583–588. - PubMed
-
- Skolnick J, Kolinski A, Ortiz AR. MONSSTER: a method for folding globular proteins with a small number of distance restraints. J. Mol. Biol. 1997;265:217–241. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

