The NADPH oxidases NOX4 and DUOX2 regulate cell cycle entry via a p53-dependent pathway
- PMID: 20531308
- PMCID: PMC2916958
- DOI: 10.1038/onc.2010.200
The NADPH oxidases NOX4 and DUOX2 regulate cell cycle entry via a p53-dependent pathway
Abstract
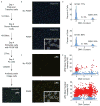
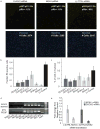
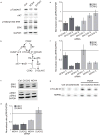
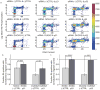
Reactive oxygen species (ROS) are produced in growth factor-signaling pathways leading to cell proliferation, but the mechanisms leading to ROS generation and the targets of ROS signals are not well understood. Using a focused siRNA screen to identify redox-related proteins required for growth factor-induced cell cycle entry, we show that two ROS-generating proteins, the NADPH oxidases NOX4 and DUOX2, are required for platelet-derived growth factor (PDGF) induced retinoblastoma protein (Rb) phosphorylation in normal human fibroblasts. Unexpectedly, NOX4 and DUOX2 knockdown did not inhibit the early signaling pathways leading to cyclin D1 upregulation. However, hours after growth factor stimulation, NOX4 and DUOX2 knockdown reduced ERK1 phosphorylation and increased levels of the tumor suppressor protein p53 and a cell cycle inhibitor protein p21 (Waf1/Cip1) that is transcriptionally regulated by p53. Co-knockdown of NOX4 or DUOX2 with either p53 or with p21 overcame the inhibition of Rb phosphorylation that occurred with NOX4 or DUOX2 knockdown alone. Our results argue that rather than primarily affecting growth factor receptor signaling, NOX4 and DUOX2 regulate cell cycle entry as part of a p53-dependent checkpoint for proliferation.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest.
Figures


Similar articles
-
Wild-type and mutant p53 differentially regulate NADPH oxidase 4 in TGF-β-mediated migration of human lung and breast epithelial cells.Br J Cancer. 2014 May 13;110(10):2569-82. doi: 10.1038/bjc.2014.165. Epub 2014 Apr 8. Br J Cancer. 2014. PMID: 24714748 Free PMC article.
-
Nox4 and Duox1/2 Mediate Redox Activation of Mesenchymal Cell Migration by PDGF.PLoS One. 2016 Apr 25;11(4):e0154157. doi: 10.1371/journal.pone.0154157. eCollection 2016. PLoS One. 2016. PMID: 27110716 Free PMC article.
-
ROS-induced ROS release orchestrated by Nox4, Nox2, and mitochondria in VEGF signaling and angiogenesis.Am J Physiol Cell Physiol. 2017 Jun 1;312(6):C749-C764. doi: 10.1152/ajpcell.00346.2016. Epub 2017 Apr 19. Am J Physiol Cell Physiol. 2017. PMID: 28424170 Free PMC article.
-
NADPH oxidase-dependent redox signaling in TGF-β-mediated fibrotic responses.Redox Biol. 2014 Jan 20;2:267-72. doi: 10.1016/j.redox.2014.01.012. eCollection 2014. Redox Biol. 2014. PMID: 24494202 Free PMC article. Review.
-
NADPH oxidase signaling and cardiac myocyte function.J Mol Cell Cardiol. 2009 Jul;47(1):15-22. doi: 10.1016/j.yjmcc.2009.04.004. Epub 2009 Apr 15. J Mol Cell Cardiol. 2009. PMID: 19374908 Review.
Cited by
-
Dosage of Dyrk1a shifts cells within a p21-cyclin D1 signaling map to control the decision to enter the cell cycle.Mol Cell. 2013 Oct 10;52(1):87-100. doi: 10.1016/j.molcel.2013.09.009. Mol Cell. 2013. PMID: 24119401 Free PMC article.
-
Induced Trf2 deletion leads to aging vascular phenotype in mice associated with arterial telomere uncapping, senescence signaling, and oxidative stress.J Mol Cell Cardiol. 2019 Feb;127:74-82. doi: 10.1016/j.yjmcc.2018.11.014. Epub 2018 Nov 29. J Mol Cell Cardiol. 2019. PMID: 30502348 Free PMC article.
-
Effects of Functionalized Fullerenes on ROS Homeostasis Determine Their Cytoprotective or Cytotoxic Properties.Nanomaterials (Basel). 2020 Jul 19;10(7):1405. doi: 10.3390/nano10071405. Nanomaterials (Basel). 2020. PMID: 32707664 Free PMC article.
-
A two-dimensional ERK-AKT signaling code for an NGF-triggered cell-fate decision.Mol Cell. 2012 Jan 27;45(2):196-209. doi: 10.1016/j.molcel.2011.11.023. Epub 2011 Dec 28. Mol Cell. 2012. PMID: 22206868 Free PMC article.
-
The proinflammatory LTB4/BLT1 signal axis confers resistance to TGF-β1-induced growth inhibition by targeting Smad3 linker region.Oncotarget. 2015 Dec 8;6(39):41650-66. doi: 10.18632/oncotarget.6146. Oncotarget. 2015. PMID: 26497676 Free PMC article.
References
-
- Bae YS, Kang SW, Seo MS, Baines IC, Tekle E, Chock PB, et al. Epidermal growth factor (EGF)-induced generation of hydrogen peroxide. Role in EGF receptor-mediated tyrosine phosphorylation. The Journal of biological chemistry. 1997;272:217–221. - PubMed
-
- Bae YS, Sung JY, Kim OS, Kim YJ, Hur KC, Kazlauskas A, et al. Platelet-derived growth factor-induced H(2)O(2) production requires the activation of phosphatidylinositol 3-kinase. The Journal of biological chemistry. 2000;275:10527–10531. - PubMed
-
- Burch PM, Heintz NH. Redox regulation of cell-cycle re-entry: cyclin D1 as a primary target for the mitogenic effects of reactive oxygen and nitrogen species. Antioxidants & redox signaling. 2005;7:741–751. - PubMed
-
- Burhans WC, Heintz NH. The cell cycle is a redox cycle: Linking phase-specific targets to cell fate. Free radical biology & medicine 2009 - PubMed
-
- Chambard JC, Lefloch R, Pouyssegur J, Lenormand P. ERK implication in cell cycle regulation. Biochimica et biophysica acta. 2007;1773:1299–1310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

