Automated identification of pathways from quantitative genetic interaction data
- PMID: 20531408
- PMCID: PMC2913392
- DOI: 10.1038/msb.2010.27
Automated identification of pathways from quantitative genetic interaction data
Abstract
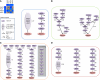
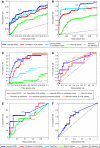
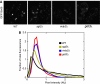
High-throughput quantitative genetic interaction (GI) measurements provide detailed information regarding the structure of the underlying biological pathways by reporting on functional dependencies between genes. However, the analytical tools for fully exploiting such information lag behind the ability to collect these data. We present a novel Bayesian learning method that uses quantitative phenotypes of double knockout organisms to automatically reconstruct detailed pathway structures. We applied our method to a recent data set that measures GIs for endoplasmic reticulum (ER) genes, using the unfolded protein response as a quantitative phenotype. The results provided reconstructions of known functional pathways including N-linked glycosylation and ER-associated protein degradation. It also contained novel relationships, such as the placement of SGT2 in the tail-anchored biogenesis pathway, a finding that we experimentally validated. Our approach should be readily applicable to the next generation of quantitative GI data sets, as assays become available for additional phenotypes and eventually higher-level organisms.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 - PMC - PubMed
-
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, Agami R, Ge W, Cavet G, Linsley PS, Beijersbergen RL, Bernards R (2004) A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428: 431–437 - PubMed
-
- Brachmann C, Davies A, Cost G, Caputo E, Li J, Hieter P, Boeke J (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 - PubMed

