Genome wide analysis of inbred mouse lines identifies a locus containing Ppar-gamma as contributing to enhanced malaria survival
- PMID: 20531941
- PMCID: PMC2878346
- DOI: 10.1371/journal.pone.0010903
Genome wide analysis of inbred mouse lines identifies a locus containing Ppar-gamma as contributing to enhanced malaria survival
Abstract
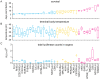
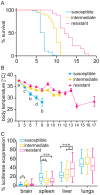
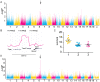
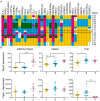
The genetic background of a patient determines in part if a person develops a mild form of malaria and recovers, or develops a severe form and dies. We have used a mouse model to detect genes involved in the resistance or susceptibility to Plasmodium berghei malaria infection. To this end we first characterized 32 different mouse strains infected with P. berghei and identified survival as the best trait to discriminate between the strains. We found a locus on chromosome 6 by linking the survival phenotypes of the mouse strains to their genetic variations using genome wide analyses such as haplotype associated mapping and the efficient mixed-model for association. This new locus involved in malaria resistance contains only two genes and confirms the importance of Ppar-gamma in malaria infection.
Conflict of interest statement
Figures
Similar articles
-
Identification of the Plasmodium berghei resistance locus 9 linked to survival on chromosome 9.Malar J. 2013 Sep 11;12:316. doi: 10.1186/1475-2875-12-316. Malar J. 2013. PMID: 24025732 Free PMC article.
-
Malaria liver stage susceptibility locus identified on mouse chromosome 17 by congenic mapping.PLoS One. 2008 Mar 26;3(3):e1874. doi: 10.1371/journal.pone.0001874. PLoS One. 2008. PMID: 18365019 Free PMC article.
-
Genetic control of parasite clearance leads to resistance to Plasmodium berghei ANKA infection and confers immunity.Genes Immun. 2005 Aug;6(5):416-21. doi: 10.1038/sj.gene.6364219. Genes Immun. 2005. PMID: 15973462
-
Genetic analysis of cerebral malaria in the mouse model infected with Plasmodium berghei.Mamm Genome. 2018 Aug;29(7-8):488-506. doi: 10.1007/s00335-018-9752-9. Epub 2018 Jun 19. Mamm Genome. 2018. PMID: 29922917 Review.
-
Complex genetic control of susceptibility to malaria in mice.Genes Immun. 2002 Jun;3(4):177-86. doi: 10.1038/sj.gene.6363841. Genes Immun. 2002. PMID: 12058252 Review.
Cited by
-
Effect of Brugia pahangi co-infection with Plasmodium berghei ANKA in gerbils (Meriones unguiculatus).Parasitol Res. 2020 Apr;119(4):1301-1315. doi: 10.1007/s00436-020-06632-4. Epub 2020 Mar 16. Parasitol Res. 2020. PMID: 32179986
-
The Case for the Use of PPARγ Agonists as an Adjunctive Therapy for Cerebral Malaria.PPAR Res. 2012;2012:513865. doi: 10.1155/2012/513865. Epub 2011 Jun 9. PPAR Res. 2012. PMID: 21772838 Free PMC article.
-
Mouse ENU Mutagenesis to Understand Immunity to Infection: Methods, Selected Examples, and Perspectives.Genes (Basel). 2014 Sep 29;5(4):887-925. doi: 10.3390/genes5040887. Genes (Basel). 2014. PMID: 25268389 Free PMC article. Review.
-
Host genetics in malaria: lessons from mouse studies.Mamm Genome. 2018 Aug;29(7-8):507-522. doi: 10.1007/s00335-018-9744-9. Epub 2018 Mar 28. Mamm Genome. 2018. PMID: 29594458 Review.
-
Genetic control of murine invariant natural killer T-cell development dynamically differs dependent on the examined tissue type.Genes Immun. 2012 Feb;13(2):164-74. doi: 10.1038/gene.2011.68. Epub 2011 Sep 22. Genes Immun. 2012. PMID: 21938016 Free PMC article.
References
-
- McGuire W, Hill AV, Allsopp CE, Greenwood BM, Kwiatkowski D. Variation in the TNF-alpha promoter region associated with susceptibility to cerebral malaria. Nature. 1994;371:508–510. - PubMed
-
- Flint J, Hill AV, Bowden DK, Oppenheimer SJ, Sill PR, et al. High frequencies of alpha-thalassaemia are the result of natural selection by malaria. Nature. 1986;321:744–750. - PubMed
-
- Ruwende C, Khoo SC, Snow RW, Yates SN, Kwiatkowski D, et al. Natural selection of hemi- and heterozygotes for G6PD deficiency in Africa by resistance to severe malaria. Nature. 1995;376:246–249. - PubMed
-
- Frodsham AJ, Hill AVS. Genetics of infectious diseases. Hum Mol Genet. 2004;13:R187–194. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

