The SOCS-box of HIV-1 Vif interacts with ElonginBC by induced-folding to recruit its Cul5-containing ubiquitin ligase complex
- PMID: 20532212
- PMCID: PMC2880568
- DOI: 10.1371/journal.ppat.1000925
The SOCS-box of HIV-1 Vif interacts with ElonginBC by induced-folding to recruit its Cul5-containing ubiquitin ligase complex
Abstract
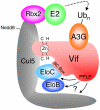
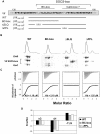
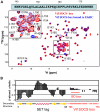
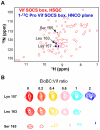
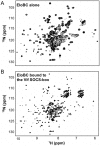
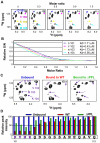
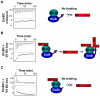
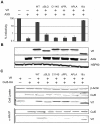
The HIV-1 viral infectivity factor (Vif) protein recruits an E3 ubiquitin ligase complex, comprising the cellular proteins elongin B and C (EloBC), cullin 5 (Cul5) and RING-box 2 (Rbx2), to the anti-viral proteins APOBEC3G (A3G) and APOBEC3F (A3F) and induces their polyubiquitination and proteasomal degradation. In this study, we used purified proteins and direct in vitro binding assays, isothermal titration calorimetry and NMR spectroscopy to describe the molecular mechanism for assembly of the Vif-EloBC ternary complex. We demonstrate that Vif binds to EloBC in two locations, and that both interactions induce structural changes in the SOCS box of Vif as well as EloBC. In particular, in addition to the previously established binding of Vif's BC box to EloC, we report a novel interaction between the conserved Pro-Pro-Leu-Pro motif of Vif and the C-terminal domain of EloB. Using cell-based assays, we further show that this interaction is necessary for the formation of a functional ligase complex, thus establishing a role of this motif. We conclude that HIV-1 Vif engages EloBC via an induced-folding mechanism that does not require additional co-factors, and speculate that these features distinguish Vif from other EloBC specificity factors such as cellular SOCS proteins, and may enhance the prospects of obtaining therapeutic inhibitors of Vif function.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. - PubMed
-
- Willems AR, Schwab M, Tyers M. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133–170. - PubMed
-
- Mahrour N, Redwine WB, Florens L, Swanson SK, Martin-Brown S, et al. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J Biol Chem. 2008;283:8005–8013. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

