Characteristics of transposable element exonization within human and mouse
- PMID: 20532223
- PMCID: PMC2879366
- DOI: 10.1371/journal.pone.0010907
Characteristics of transposable element exonization within human and mouse
Abstract
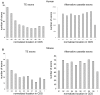
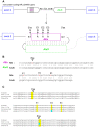
Insertion of transposed elements within mammalian genes is thought to be an important contributor to mammalian evolution and speciation. Insertion of transposed elements into introns can lead to their activation as alternatively spliced cassette exons, an event called exonization. Elucidation of the evolutionary constraints that have shaped fixation of transposed elements within human and mouse protein coding genes and subsequent exonization is important for understanding of how the exonization process has affected transcriptome and proteome complexities. Here we show that exonization of transposed elements is biased towards the beginning of the coding sequence in both human and mouse genes. Analysis of single nucleotide polymorphisms (SNPs) revealed that exonization of transposed elements can be population-specific, implying that exonizations may enhance divergence and lead to speciation. SNP density analysis revealed differences between Alu and other transposed elements. Finally, we identified cases of primate-specific Alu elements that depend on RNA editing for their exonization. These results shed light on TE fixation and the exonization process within human and mouse genes.
Conflict of interest statement
Figures
Similar articles
-
Biased exonization of transposed elements in duplicated genes: A lesson from the TIF-IA gene.BMC Mol Biol. 2007 Nov 29;8:109. doi: 10.1186/1471-2199-8-109. BMC Mol Biol. 2007. PMID: 18047649 Free PMC article.
-
SERpredict: detection of tissue- or tumor-specific isoforms generated through exonization of transposable elements.BMC Genet. 2007 Nov 6;8:78. doi: 10.1186/1471-2156-8-78. BMC Genet. 2007. PMID: 17986331 Free PMC article.
-
Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome.Genome Biol. 2007;8(6):R127. doi: 10.1186/gb-2007-8-6-r127. Genome Biol. 2007. PMID: 17594509 Free PMC article.
-
Exonization of transposed elements: A challenge and opportunity for evolution.Biochimie. 2011 Nov;93(11):1928-34. doi: 10.1016/j.biochi.2011.07.014. Epub 2011 Jul 26. Biochimie. 2011. PMID: 21787833 Review.
-
The role of Alu elements in the cis-regulation of RNA processing.Cell Mol Life Sci. 2015 Nov;72(21):4063-76. doi: 10.1007/s00018-015-1990-3. Epub 2015 Jul 30. Cell Mol Life Sci. 2015. PMID: 26223268 Free PMC article. Review.
Cited by
-
Eusociality in snapping shrimps is associated with larger genomes and an accumulation of transposable elements.Proc Natl Acad Sci U S A. 2021 Jun 15;118(24):e2025051118. doi: 10.1073/pnas.2025051118. Proc Natl Acad Sci U S A. 2021. PMID: 34099551 Free PMC article.
-
Integrative genomics and transcriptomics analysis of human embryonic and induced pluripotent stem cells.BioData Min. 2014 Dec 13;7(1):32. doi: 10.1186/s13040-014-0032-2. eCollection 2014. BioData Min. 2014. PMID: 25649046 Free PMC article.
-
Perspectives on studying molecular adaptations of amphibians in the genomic era.Zool Res. 2020 Jul 18;41(4):351-364. doi: 10.24272/j.issn.2095-8137.2020.046. Zool Res. 2020. PMID: 32390371 Free PMC article. Review.
-
Inferring the expression variability of human transposable element-derived exons by linear model analysis of deep RNA sequencing data.BMC Genomics. 2013 Aug 28;14:584. doi: 10.1186/1471-2164-14-584. BMC Genomics. 2013. PMID: 23984937 Free PMC article.
-
RNA sequencing of the exercise transcriptome in equine athletes.PLoS One. 2013 Dec 31;8(12):e83504. doi: 10.1371/journal.pone.0083504. eCollection 2013. PLoS One. 2013. PMID: 24391776 Free PMC article.
References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. - PubMed
-
- Sironi M, Menozzi G, Comi GP, Bresolin N, Cagliani R, et al. Fixation of conserved sequences shapes human intron size and influences transposon-insertion dynamics. Trends Genet. 2005;21:484–488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

