Analysis of PKR activation using analytical ultracentrifugation
- PMID: 20533534
- PMCID: PMC2926283
- DOI: 10.1002/mabi.201000069
Analysis of PKR activation using analytical ultracentrifugation
Abstract
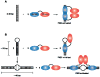
Protein kinase R (PKR) is a central component of the interferon antiviral defense pathway. Upon binding to dsRNA, PKR undergoes autophosphorylation reactions that activate the kinase, resulting in the inhibition of protein synthesis in virally-infected cells. We have used analytical ultracentrifugation and related biophysical methods to quantitatively characterize the stoichiometries, affinities, and free energy couplings that govern the assembly of the macromolecular complexes in the PKR activation pathway. These studies demonstrate that PKR dimerization play a key role in enzymatic activation and support a model where the role of dsRNA is to bring two or more PKR monomers in close proximity to enhance dimerization.
Figures

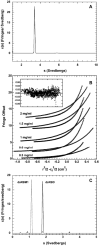
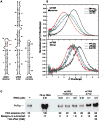
 ) and 24,000 (□) RPM; temperature, 20°C; interference optics. The dataset was globally fit to monomer-dimer model yielding Kd= 446 μM. The solid lines show the best-fit model and the inset shows the residuals. C) Sedimentation velocity analysis of PKR dsRNA binding domain constructs.[35] Continuous sedimentation coefficient distribution analysis of dsRBD (
) and 24,000 (□) RPM; temperature, 20°C; interference optics. The dataset was globally fit to monomer-dimer model yielding Kd= 446 μM. The solid lines show the best-fit model and the inset shows the residuals. C) Sedimentation velocity analysis of PKR dsRNA binding domain constructs.[35] Continuous sedimentation coefficient distribution analysis of dsRBD ( ) and dsRBM 1 (
) and dsRBM 1 ( ). Conditions: sample concentration, 0.5 mg/ml (dsRBD) and 0.6 mg/ml (dsRBM 1); Rotor speed, 50,000 RPM; temperature, 20°C; interference optics.
). Conditions: sample concentration, 0.5 mg/ml (dsRBD) and 0.6 mg/ml (dsRBM 1); Rotor speed, 50,000 RPM; temperature, 20°C; interference optics.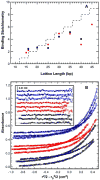
 ), 260 nm (□) and 280 nm (△). Solid lines are a global fit of the data to an unconstrained model of three ligands binding to the 20 mer RNA yielding Kd1= 11 nM, Kd2 = 210 nM and Kd3= 780 nM and an RMSD = 0.00437 OD. Inset: residuals. Traces have been vertically offset for clarity.
), 260 nm (□) and 280 nm (△). Solid lines are a global fit of the data to an unconstrained model of three ligands binding to the 20 mer RNA yielding Kd1= 11 nM, Kd2 = 210 nM and Kd3= 780 nM and an RMSD = 0.00437 OD. Inset: residuals. Traces have been vertically offset for clarity.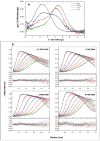

Similar articles
-
Analysis of high-affinity binding of protein kinase R to double-stranded RNA.Biochemistry. 2012 Nov 6;51(44):8764-70. doi: 10.1021/bi301226h. Epub 2012 Oct 26. Biochemistry. 2012. PMID: 23062027 Free PMC article.
-
Activation of PKR by short stem-loop RNAs containing single-stranded arms.RNA. 2016 Jul;22(7):1065-75. doi: 10.1261/rna.053348.115. Epub 2016 May 20. RNA. 2016. PMID: 27208315 Free PMC article.
-
Mechanism of PKR Activation by dsRNA.J Mol Biol. 2008 Aug 29;381(2):351-60. doi: 10.1016/j.jmb.2008.05.056. Epub 2008 May 29. J Mol Biol. 2008. PMID: 18599071 Free PMC article.
-
Activation of PKR: an open and shut case?Trends Biochem Sci. 2007 Feb;32(2):57-62. doi: 10.1016/j.tibs.2006.12.003. Epub 2006 Dec 29. Trends Biochem Sci. 2007. PMID: 17196820 Free PMC article. Review.
-
The regulation of the protein kinase PKR by RNA.Biochimie. 1996;78(11-12):909-14. doi: 10.1016/s0300-9084(97)86712-0. Biochimie. 1996. PMID: 9150867 Review.
Cited by
-
Auto-phosphorylation Represses Protein Kinase R Activity.Sci Rep. 2017 Mar 10;7:44340. doi: 10.1038/srep44340. Sci Rep. 2017. PMID: 28281686 Free PMC article.
-
Heparin activates PKR by inducing dimerization.J Mol Biol. 2011 Nov 11;413(5):973-84. doi: 10.1016/j.jmb.2011.09.025. Epub 2011 Sep 28. J Mol Biol. 2011. PMID: 21978664 Free PMC article.
-
Protein-protein interactions: switch from classical methods to proteomics and bioinformatics-based approaches.Cell Mol Life Sci. 2014 Jan;71(2):205-28. doi: 10.1007/s00018-013-1333-1. Epub 2013 Apr 12. Cell Mol Life Sci. 2014. PMID: 23579629 Free PMC article. Review.
-
Regulation of innate immunity through RNA structure and the protein kinase PKR.Curr Opin Struct Biol. 2011 Feb;21(1):119-27. doi: 10.1016/j.sbi.2010.11.003. Epub 2010 Dec 8. Curr Opin Struct Biol. 2011. PMID: 21145228 Free PMC article. Review.
-
Multi-level regulation of cellular recognition of viral dsRNA.Cell Mol Life Sci. 2013 Jun;70(11):1949-63. doi: 10.1007/s00018-012-1149-4. Epub 2012 Sep 9. Cell Mol Life Sci. 2013. PMID: 22960755 Free PMC article. Review.
References
-
- Kaufman RJ. The double stranded RNA-activated protein kinase PKR. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. p. 503.
-
- Toth AM, Zhang P, Das S, George CX, Samuel CE. Prog Nucleic Acid Res Mol Biol. 2006;81:369. - PubMed
-
- Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. Nature Rev Mol Cell Biol. 2004;5:1013. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

