Quantitative 3D imaging of whole, unstained cells by using X-ray diffraction microscopy
- PMID: 20534442
- PMCID: PMC2895086
- DOI: 10.1073/pnas.1000156107
Quantitative 3D imaging of whole, unstained cells by using X-ray diffraction microscopy
Abstract
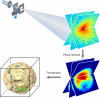
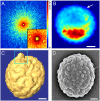
Microscopy has greatly advanced our understanding of biology. Although significant progress has recently been made in optical microscopy to break the diffraction-limit barrier, reliance of such techniques on fluorescent labeling technologies prohibits quantitative 3D imaging of the entire contents of cells. Cryoelectron microscopy can image pleomorphic structures at a resolution of 3-5 nm, but is only applicable to thin or sectioned specimens. Here, we report quantitative 3D imaging of a whole, unstained cell at a resolution of 50-60 nm by X-ray diffraction microscopy. We identified the 3D morphology and structure of cellular organelles including cell wall, vacuole, endoplasmic reticulum, mitochondria, granules, nucleus, and nucleolus inside a yeast spore cell. Furthermore, we observed a 3D structure protruding from the reconstructed yeast spore, suggesting the spore germination process. Using cryogenic technologies, a 3D resolution of 5-10 nm should be achievable by X-ray diffraction microscopy. This work hence paves a way for quantitative 3D imaging of a wide range of biological specimens at nanometer-scale resolutions that are too thick for electron microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Miao J, Charalambous P, Kirz J, Sayre D. Extending the methodology of X-ray crystallography to allow imaging of micrometre-sized non-crystalline specimens. Nature. 1999;400:342–344.
-
- Robinson IK, Vartanyants IA, Williams GJ, Pfeifer MA, Pitney JA. Reconstruction of the shapes of gold nanocrystals using coherent X-ray diffraction. Phys Rev Lett. 2001;87:195505. - PubMed
-
- Miao J, et al. High resolution 3D X-ray diffraction microscopy. Phys Rev Lett. 2002;89:088303. - PubMed
-
- Williams GJ, Pfeifer MA, Vartanyants IA, Robinson IK. Three-dimensional imaging of microstructure in Au nanocrystals. Phys Rev Lett. 2003;90:175501. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

