Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition
- PMID: 20534443
- PMCID: PMC2900659
- DOI: 10.1073/pnas.1003817107
Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition
Abstract
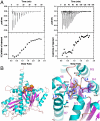
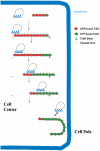
The segregation of plasmid DNA typically requires three elements: a DNA centromere site, an NTPase, and a centromere-binding protein. Because of their simplicity, plasmid partition systems represent tractable models to study the molecular basis of DNA segregation. Unlike eukaryotes, which utilize the GTPase tubulin to segregate DNA, the most common plasmid-encoded NTPases contain Walker-box and actin-like folds. Recently, a plasmid stability cassette on Bacillus thuringiensis pBtoxis encoding a putative FtsZ/tubulin-like NTPase called TubZ and DNA-binding protein called TubR has been described. How these proteins collaborate to impart plasmid stability, however, is unknown. Here we show that the TubR structure consists of an intertwined dimer with a winged helix-turn-helix (HTH) motif. Strikingly, however, the TubR recognition helices mediate dimerization, making canonical HTH-DNA interactions impossible. Mutagenesis data indicate that a basic patch, encompassing the two wing regions and the N termini of the recognition helices, mediates DNA binding, which indicates an unusual HTH-DNA interaction mode in which the N termini of the recognition helices insert into a single DNA groove and the wings into adjacent DNA grooves. The TubZ structure shows that it is as similar structurally to eukaryotic tubulin as it is to bacterial FtsZ. TubZ forms polymers with guanine nucleotide-binding characteristics and polymer dynamics similar to tubulin. Finally, we show that the exposed TubZ C-terminal region interacts with TubR-DNA, linking the TubR-bound pBtoxis to TubZ polymerization. The combined data suggest a mechanism for TubZ-polymer powered plasmid movement.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
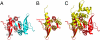
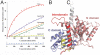


Comment in
-
One-way ticket to the cell pole: plasmid transport by the prokaryotic tubulin homolog TubZ.Proc Natl Acad Sci U S A. 2010 Jul 6;107(27):12061-2. doi: 10.1073/pnas.1007331107. Epub 2010 Jun 28. Proc Natl Acad Sci U S A. 2010. PMID: 20616084 Free PMC article. No abstract available.
Similar articles
-
Filament formation of the FtsZ/tubulin-like protein TubZ from the Bacillus cereus pXO1 plasmid.J Biol Chem. 2012 Sep 14;287(38):32103-12. doi: 10.1074/jbc.M112.373803. Epub 2012 Jul 30. J Biol Chem. 2012. PMID: 22847006 Free PMC article.
-
Cooperative DNA Binding of the Plasmid Partitioning Protein TubR from the Bacillus cereus pXO1 Plasmid.J Mol Biol. 2018 Dec 7;430(24):5015-5028. doi: 10.1016/j.jmb.2018.11.001. Epub 2018 Nov 8. J Mol Biol. 2018. PMID: 30414406
-
Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis.Genes Dev. 2007 Jun 1;21(11):1340-52. doi: 10.1101/gad.1546107. Epub 2007 May 17. Genes Dev. 2007. PMID: 17510284 Free PMC article.
-
Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation.Biochem J. 2008 May 15;412(1):1-18. doi: 10.1042/BJ20080359. Biochem J. 2008. PMID: 18426389 Review.
-
Tubulin-Like Proteins in Prokaryotic DNA Positioning.Subcell Biochem. 2017;84:323-356. doi: 10.1007/978-3-319-53047-5_11. Subcell Biochem. 2017. PMID: 28500531 Review.
Cited by
-
Structures of maintenance of carboxysome distribution Walker-box McdA and McdB adaptor homologs.Nucleic Acids Res. 2019 Jun 20;47(11):5950-5962. doi: 10.1093/nar/gkz314. Nucleic Acids Res. 2019. PMID: 31106331 Free PMC article.
-
A novel transcriptional activator, tubX, is required for the stability of Bacillus sphaericus mosquitocidal plasmid pBsph.J Bacteriol. 2014 Dec;196(24):4304-14. doi: 10.1128/JB.01855-14. Epub 2014 Sep 29. J Bacteriol. 2014. PMID: 25266379 Free PMC article.
-
Reconstitution of a prokaryotic minus end-tracking system using TubRC centromeric complexes and tubulin-like protein TubZ filaments.Proc Natl Acad Sci U S A. 2015 Apr 14;112(15):E1845-50. doi: 10.1073/pnas.1423746112. Epub 2015 Mar 30. Proc Natl Acad Sci U S A. 2015. PMID: 25825718 Free PMC article.
-
Microtubules in bacteria: Ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton.PLoS Biol. 2011 Dec;9(12):e1001213. doi: 10.1371/journal.pbio.1001213. Epub 2011 Dec 6. PLoS Biol. 2011. PMID: 22162949 Free PMC article.
-
The wing of a winged helix-turn-helix transcription factor organizes the active site of BirA, a bifunctional repressor/ligase.J Biol Chem. 2013 Dec 13;288(50):36029-39. doi: 10.1074/jbc.M113.525618. Epub 2013 Nov 4. J Biol Chem. 2013. PMID: 24189073 Free PMC article.
References
-
- Nogales E, Wolf SG, Downing KH. Structure of the αβ tubulin dimer by electron crystallography. Nature. 1998;391:199–203. - PubMed
-
- Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96:79–88. - PubMed
-
- Nogales E. Structural insights into microtubule function. Annu Rev Biochem. 2000;69:277–302. - PubMed
-
- Oliva MA, Cordell SC, Löwe J. Structural insights into FtsZ protofilament formation. Nat Struct Mol Biol. 2004;11:1243–1250. - PubMed
-
- Oliva MA, Trambaiolo D, Löwe J. Structural insights into the conformational variability of FtsZ. J Mol Biol. 2007;373:1229–1242. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

