Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase
- PMID: 20534465
- PMCID: PMC2890747
- DOI: 10.1073/pnas.1003653107
Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase
Abstract
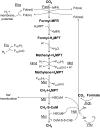
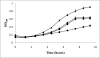
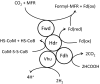
In methanogenic Archaea, the final step of methanogenesis generates methane and a heterodisulfide of coenzyme M and coenzyme B (CoM-S-S-CoB). Reduction of this heterodisulfide by heterodisulfide reductase to regenerate HS-CoM and HS-CoB is an exergonic process. Thauer et al. [Thauer, et al. 2008 Nat Rev Microbiol 6:579-591] recently suggested that in hydrogenotrophic methanogens the energy of heterodisulfide reduction powers the most endergonic reaction in the pathway, catalyzed by the formylmethanofuran dehydrogenase, via flavin-based electron bifurcation. Here we present evidence that these two steps in methanogenesis are physically linked. We identify a protein complex from the hydrogenotrophic methanogen, Methanococcus maripaludis, that contains heterodisulfide reductase, formylmethanofuran dehydrogenase, F(420)-nonreducing hydrogenase, and formate dehydrogenase. In addition to establishing a physical basis for the electron-bifurcation model of energy conservation, the composition of the complex also suggests that either H(2) or formate (two alternative electron donors for methanogenesis) can donate electrons to the heterodisulfide-H(2) via F(420)-nonreducing hydrogenase or formate via formate dehydrogenase. Electron flow from formate to the heterodisulfide rather than the use of H(2) as an intermediate represents a previously unknown path of electron flow in methanogenesis. We further tested whether this path occurs by constructing a mutant lacking F(420)-nonreducing hydrogenase. The mutant displayed growth equal to wild-type with formate but markedly slower growth with hydrogen. The results support the model of electron bifurcation and suggest that formate, like H(2), is closely integrated into the methanogenic pathway.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic Archaea: Ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–591. - PubMed
-
- Tumbula DL, Whitman WB. Genetics of Methanococcus: Possibilities for functional genomics in Archaea. Mol Microbiol. 1999;33:1–7. - PubMed
-
- Haydock AK, Porat I, Whitman WB, Leigh JA. Continuous culture of Methanococcus maripaludis under defined nutrient conditions. FEMS Microbiol Lett. 2004;238:85–91. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

