Influenza A virus-generated small RNAs regulate the switch from transcription to replication
- PMID: 20534471
- PMCID: PMC2895093
- DOI: 10.1073/pnas.1001984107
Influenza A virus-generated small RNAs regulate the switch from transcription to replication
Abstract
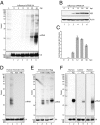
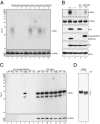
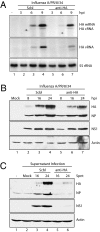
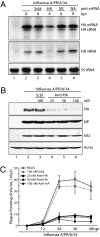
The discovery of regulatory small RNAs continues to reshape paradigms in both molecular biology and virology. Here we describe examples of influenza A virus-derived small viral RNAs (svRNAs). svRNAs are 22-27 nt in length and correspond to the 5' end of each of the viral genomic RNA (vRNA) segments. Expression of svRNA correlates with the accumulation of vRNA and a bias in RNA-dependent RNA polymerase (RdRp) activity from transcription toward genome replication. Synthesis of svRNA requires the RdRp, nucleoprotein and the nuclear export protein NS2. In addition, svRNA is detectable during replication of various influenza A virus subtypes across multiple host species and associates physically with the RdRp. We demonstrate that depletion of svRNA has a minimal impact on mRNA and complementary vRNA (cRNA) but results in a dramatic loss of vRNA in a segment-specific manner. We propose that svRNA triggers the viral switch from transcription to replication through interactions with the viral polymerase machinery. Taken together, the discovery of svRNA redefines the mechanistic switch of influenza virus transcription/replication and provides a potential target for broad-range, anti-influenza virus-based therapeutics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
A big role for small RNAs in influenza virus replication.Proc Natl Acad Sci U S A. 2010 Jun 22;107(25):11153-4. doi: 10.1073/pnas.1006673107. Epub 2010 Jun 14. Proc Natl Acad Sci U S A. 2010. PMID: 20547839 Free PMC article. No abstract available.
References
-
- Palese P, Shaw M. In: Fields Virology. 5th Ed. Knipe DM, Howley PM, editors. Philadelphia: Raven; 2007. pp. 1648–1698.
-
- Krug RM. Priming of influenza viral RNA transcription by capped heterologous RNAs. Curr Top Microbiol Immunol. 1981;93:125–149. - PubMed
-
- Tarendeau F, et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat Struct Mol Biol. 2007;14:229–233. - PubMed
-
- Guilligay D, et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat Struct Mol Biol. 2008;15:500–506. - PubMed
-
- He X, et al. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature. 2008;454:1123–1126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

