Crystal structure of soluble MD-1 and its interaction with lipid IVa
- PMID: 20534476
- PMCID: PMC2890730
- DOI: 10.1073/pnas.1004153107
Crystal structure of soluble MD-1 and its interaction with lipid IVa
Abstract
Lipopolysaccharide (LPS) of Gram-negative bacteria is a common pathogen-associated molecular pattern (PAMP) that induces potent innate immune responses. The host immune response against LPS is triggered by myeloid differentiation factor 2 (MD-2) in association with Toll-like receptor 4 (TLR4) on the cell surface. The MD-2/TLR4-mediated LPS response is regulated by the evolutionarily related complex of MD-1 and Toll-like receptor homolog RP105. Here, we report crystallographic and biophysical data that demonstrate a previously unidentified direct interaction of MD-1 with LPS. The crystal structure of chicken MD-1 (cMD-1) at 2.0 A resolution exhibits a beta-cup-like fold, similar to MD-2, that encloses a hydrophobic cavity between the two beta-sheets. A lipid-like moiety was observed inside the cavity, suggesting the possibility of a direct MD-1/LPS interaction. LPS was subsequently identified as an MD-1 ligand by native gel electrophoresis and gel filtration analyses. The crystal structure of cMD-1 with lipid IVa, an LPS precursor, at 2.4 A resolution revealed that the lipid inserts into the deep hydrophobic cavity of the beta-cup-like structure, but with some important differences compared with MD-2. These findings suggest that soluble MD-1 alone, in addition to its complex with RP105, can regulate host LPS sensitivity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
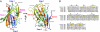




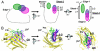
References
-
- Gangloff M, Gay NJ. MD-2: The Toll ‘gatekeeper’ in endotoxin signalling. Trends Biochem Sci. 2004;29:294–300. - PubMed
-
- Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;348:1546–1554. - PubMed
-
- Beutler B, Rietschel ET. Innate immune sensing and its roots: The story of endotoxin. Nat Rev Immunol. 2003;3:169–176. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

