Adaptive impact of the chimeric gene Quetzalcoatl in Drosophila melanogaster
- PMID: 20534482
- PMCID: PMC2890713
- DOI: 10.1073/pnas.1006503107
Adaptive impact of the chimeric gene Quetzalcoatl in Drosophila melanogaster
Abstract
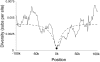
Chimeric genes, which form through the genomic fusion of two protein-coding genes, are a significant source of evolutionary novelty in Drosophila melanogaster. However, the propensity of chimeric genes to produce adaptive phenotypic changes is not fully understood. Here, we describe the chimeric gene Quetzalcoatl (Qtzl; CG31864), which formed in the recent past and swept to fixation in D. melanogaster. Qtzl arose through a duplication on chromosome 2L that united a portion of the mitochondrially targeted peptide CG12264 with a segment of the polycomb gene escl. The 3' segment of the gene, which is derived from escl, is inherited out of frame, producing a unique peptide sequence. Nucleotide diversity is drastically reduced and site frequency spectra are significantly skewed surrounding the duplicated region, a finding consistent with a selective sweep on the duplicate region containing Qtzl. Qtzl has an expression profile that largely resembles that of escl, with expression in early pupae, adult females, and male testes. However, expression patterns appear to have been decoupled from both parental genes during later embryonic development and in head tissues of adult males, indicating that Qtzl has developed a distinct regulatory profile through the rearrangement of different 5' and 3' regulatory domains. Furthermore, misexpression of Qtzl suppresses defects in the formation of the neuromuscular junction in larvae, demonstrating that Qtzl can produce phenotypic effects in cells. Together, these results show that chimeric genes can produce structural and regulatory changes in a single mutational step and may be a major factor in adaptive evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
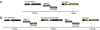
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

