Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins
- PMID: 20534494
- PMCID: PMC2890729
- DOI: 10.1073/pnas.1003015107
Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins
Abstract
The RNA-dependent RNA polymerase core complex formed upon infection of Escherichia coli by the bacteriophage Qbeta is composed of the viral catalytic beta-subunit as well as the host translation elongation factors EF-Tu and EF-Ts, which are required for initiation of RNA replication. We have determined the crystal structure of the complex between the beta-subunit and the two host proteins to 2.5-A resolution. Whereas the basic catalytic machinery in the viral subunit appears similar to other RNA-dependent RNA polymerases, a unique C-terminal region of the beta-subunit engages in extensive interactions with EF-Tu and may contribute to the separation of the transient duplex formed between the template and the nascent product to allow exponential amplification of the phage genome. The evolution of resistance by the host appears to be impaired because of the interactions of the beta-subunit with parts of EF-Tu essential in recognition of aminoacyl-tRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
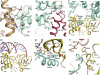
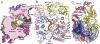
Similar articles
-
Structural basis for RNA-genome recognition during bacteriophage Qβ replication.Nucleic Acids Res. 2015 Dec 15;43(22):10893-906. doi: 10.1093/nar/gkv1212. Epub 2015 Nov 17. Nucleic Acids Res. 2015. PMID: 26578560 Free PMC article.
-
Assembly of Q{beta} viral RNA polymerase with host translational elongation factors EF-Tu and -Ts.Proc Natl Acad Sci U S A. 2010 Sep 7;107(36):15733-8. doi: 10.1073/pnas.1006559107. Epub 2010 Aug 23. Proc Natl Acad Sci U S A. 2010. PMID: 20798060 Free PMC article.
-
Structures and functions of Qβ replicase: translation factors beyond protein synthesis.Int J Mol Sci. 2014 Sep 2;15(9):15552-70. doi: 10.3390/ijms150915552. Int J Mol Sci. 2014. PMID: 25184952 Free PMC article. Review.
-
Molecular basis for RNA polymerization by Qβ replicase.Nat Struct Mol Biol. 2012 Jan 15;19(2):229-37. doi: 10.1038/nsmb.2204. Nat Struct Mol Biol. 2012. PMID: 22245970
-
[Paradoxes of replication of RNA of a bacterial virus].Mol Biol (Mosk). 2011 Jan-Feb;45(1):160-72. Mol Biol (Mosk). 2011. PMID: 21485505 Review. Russian.
Cited by
-
A design principle for a single-stranded RNA genome that replicates with less double-strand formation.Nucleic Acids Res. 2015 Sep 18;43(16):8033-43. doi: 10.1093/nar/gkv742. Epub 2015 Jul 21. Nucleic Acids Res. 2015. PMID: 26202975 Free PMC article.
-
Experimental selection reveals a trade-off between fecundity and lifespan in the coliphage Qß.Open Biol. 2013 Jun 12;3(6):130043. doi: 10.1098/rsob.130043. Open Biol. 2013. PMID: 23760365 Free PMC article.
-
Structural basis for RNA-genome recognition during bacteriophage Qβ replication.Nucleic Acids Res. 2015 Dec 15;43(22):10893-906. doi: 10.1093/nar/gkv1212. Epub 2015 Nov 17. Nucleic Acids Res. 2015. PMID: 26578560 Free PMC article.
-
The dependence of viral RNA replication on co-opted host factors.Nat Rev Microbiol. 2011 Dec 19;10(2):137-49. doi: 10.1038/nrmicro2692. Nat Rev Microbiol. 2011. PMID: 22183253 Free PMC article. Review.
-
NMR structure of the Vibrio vulnificus ribosomal protein S1 domains D3 and D4 provides insights into molecular recognition of single-stranded RNAs.Nucleic Acids Res. 2021 Jul 21;49(13):7753-7764. doi: 10.1093/nar/gkab562. Nucleic Acids Res. 2021. PMID: 34223902 Free PMC article.
References
-
- Su Q, Schuppli D, Tsui HCT, Winkler ME, Weber H. Strongly reduced phage Qβ replication, but normal phage MS2 replication in an Escherichia coli K12 mutant with inactivated Qβ host factor (hfq) gene. Virology. 1997;227:211–214. - PubMed
-
- Munishkin AV, et al. Efficient templates for Qβ replicase are formed by recombination from heterologous sequences. J Mol Biol. 1991;221:463–472. - PubMed
-
- Kamen R, Kondo M, Römer W, Weissmann C. Reconstitution of Qβ replicase lacking subunit with protein-synthesis-interference factor i. Eur J Biochem. 1972;31:44–51. - PubMed
-
- Weissmann C. The making of a phage. FEBS Lett. 1974;40:S10–18. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

