Protein interface conservation across structure space
- PMID: 20534496
- PMCID: PMC2890749
- DOI: 10.1073/pnas.1005894107
Protein interface conservation across structure space
Abstract
With the advent of Systems Biology, the prediction of whether two proteins form a complex has become a problem of increased importance. A variety of experimental techniques have been applied to the problem, but three-dimensional structural information has not been widely exploited. Here we explore the range of applicability of such information by analyzing the extent to which the location of binding sites on protein surfaces is conserved among structural neighbors. We find, as expected, that interface conservation is most significant among proteins that have a clear evolutionary relationship, but that there is a significant level of conservation even among remote structural neighbors. This finding is consistent with recent evidence that information available from structural neighbors, independent of classification, should be exploited in the search for functional insights. The value of such structural information is highlighted through the development of a new protein interface prediction method, PredUs, that identifies what residues on protein surfaces are likely to participate in complexes with other proteins. The performance of PredUs, as measured through comparisons with other methods, suggests that relationships across protein structure space can be successfully exploited in the prediction of protein-protein interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
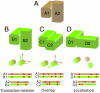

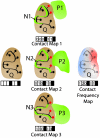
 of an atom of a surface residue of Q after applying the transformation, that residue is marked (red circles), generating a contact map for each structural neighbor (black boxes represent nonsurface residues that are not included). The “contact frequency map” is generated by summing the individual contact maps.
of an atom of a surface residue of Q after applying the transformation, that residue is marked (red circles), generating a contact map for each structural neighbor (black boxes represent nonsurface residues that are not included). The “contact frequency map” is generated by summing the individual contact maps.References
-
- Salwinski L, Eisenberg D. Computational methods of analysis of protein-protein interactions. Curr Opin Struct Biol. 2003;13:377–382. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

