Protein folded states are kinetic hubs
- PMID: 20534497
- PMCID: PMC2890711
- DOI: 10.1073/pnas.1003962107
Protein folded states are kinetic hubs
Erratum in
- Proc Natl Acad Sci U S A. 2010 Sep 21;107(38):16749
Abstract
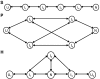
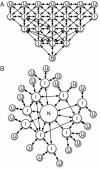
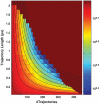
Understanding molecular kinetics, and particularly protein folding, is a classic grand challenge in molecular biophysics. Network models, such as Markov state models (MSMs), are one potential solution to this problem. MSMs have recently yielded quantitative agreement with experimentally derived structures and folding rates for specific systems, leaving them positioned to potentially provide a deeper understanding of molecular kinetics that can lead to experimentally testable hypotheses. Here we use existing MSMs for the villin headpiece and NTL9, which were constructed from atomistic simulations, to accomplish this goal. In addition, we provide simpler, humanly comprehensible networks that capture the essence of molecular kinetics and reproduce qualitative phenomena like the apparent two-state folding often seen in experiments. Together, these models show that protein dynamics are dominated by stochastic jumps between numerous metastable states and that proteins have heterogeneous unfolded states (many unfolded basins that interconvert more rapidly with the native state than with one another) yet often still appear two-state. Most importantly, we find that protein native states are hubs that can be reached quickly from any other state. However, metastability and a web of nonnative states slow the average folding rate. Experimental tests for these findings and their implications for other fields, like protein design, are also discussed.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Uversky VN. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front Biosci. 2009;14:5188–5238. - PubMed
-
- Schutte C. Berlin: Freie Universitat; 1999. Conformational dynamics: Modeling, theory, algorithm, and application to biomolecules. PhD thesis.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

