Central role of the RNA polymerase trigger loop in intrinsic RNA hydrolysis
- PMID: 20534498
- PMCID: PMC2890756
- DOI: 10.1073/pnas.0914424107
Central role of the RNA polymerase trigger loop in intrinsic RNA hydrolysis
Abstract
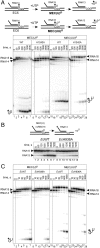
The active center of RNA polymerase can hydrolyze phosphodiester bonds in nascent RNA, a reaction thought to be important for proofreading of transcription. The reaction proceeds via a general two Mg(2+) mechanism and is assisted by the 3' end nucleotide of the transcript. Here, by using Thermus aquaticus RNA polymerase, we show that the reaction also requires the flexible domain of the active center, the trigger loop (TL). We show that the invariant histidine (beta' His1242) of the TL is essential for hydrolysis/proofreading and participates in the reaction in two distinct ways: by positioning the 3' end nucleotide of the transcript that assists catalysis and/or by directly participating in the reaction as a general base. We also show that participation of the beta' His1242 of the TL in phosphodiester bond hydrolysis does not depend on the extent of elongation complex backtracking. We obtained similar results with Escherichia coli RNA polymerase, indicating that the function of the TL in phosphodiester bond hydrolysis is conserved among bacteria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Two distinct pathways of RNA polymerase backtracking determine the requirement for the Trigger Loop during RNA hydrolysis.Nucleic Acids Res. 2021 Sep 7;49(15):8777-8784. doi: 10.1093/nar/gkab675. Nucleic Acids Res. 2021. PMID: 34365509 Free PMC article.
-
Trigger loop of RNA polymerase is a positional, not acid-base, catalyst for both transcription and proofreading.Proc Natl Acad Sci U S A. 2017 Jun 27;114(26):E5103-E5112. doi: 10.1073/pnas.1702383114. Epub 2017 Jun 12. Proc Natl Acad Sci U S A. 2017. PMID: 28607053 Free PMC article.
-
Mutations in RNA Polymerase Bridge Helix and Switch Regions Affect Active-Site Networks and Transcript-Assisted Hydrolysis.J Mol Biol. 2015 Nov 6;427(22):3516-3526. doi: 10.1016/j.jmb.2015.09.005. Epub 2015 Sep 10. J Mol Biol. 2015. PMID: 26365052 Free PMC article.
-
RNA polymerase structure-function: insights into points of transcriptional regulation.Curr Opin Microbiol. 2000 Apr;3(2):118-25. doi: 10.1016/s1369-5274(00)00062-x. Curr Opin Microbiol. 2000. PMID: 10744988 Review.
-
Mechanisms of Bacterial Transcription Termination: All Good Things Must End.Annu Rev Biochem. 2016 Jun 2;85:319-47. doi: 10.1146/annurev-biochem-060815-014844. Epub 2016 Mar 17. Annu Rev Biochem. 2016. PMID: 27023849 Review.
Cited by
-
Activation and reactivation of the RNA polymerase II trigger loop for intrinsic RNA cleavage and catalysis.Transcription. 2014;5(3):e28869. doi: 10.4161/trns.28869. Transcription. 2014. PMID: 25764335 Free PMC article.
-
Intrinsic Cleavage of RNA Polymerase II Adopts a Nucleobase-independent Mechanism Assisted by Transcript Phosphate.Nat Energy. 2019 Mar;2(3):228-235. doi: 10.1038/s41929-019-0227-5. Epub 2019 Feb 11. Nat Energy. 2019. PMID: 31179024 Free PMC article.
-
Lineage-specific variations in the trigger loop modulate RNA proofreading by bacterial RNA polymerases.Nucleic Acids Res. 2016 Feb 18;44(3):1298-308. doi: 10.1093/nar/gkv1521. Epub 2016 Jan 4. Nucleic Acids Res. 2016. PMID: 26733581 Free PMC article.
-
Transcript assisted phosphodiester bond hydrolysis by eukaryotic RNA polymerase II.Transcription. 2013 Sep-Dec;4(5):209-12. doi: 10.4161/trns.27062. Transcription. 2013. PMID: 24270513 Free PMC article.
-
Two distinct pathways of RNA polymerase backtracking determine the requirement for the Trigger Loop during RNA hydrolysis.Nucleic Acids Res. 2021 Sep 7;49(15):8777-8784. doi: 10.1093/nar/gkab675. Nucleic Acids Res. 2021. PMID: 34365509 Free PMC article.
References
-
- Steitz TA. A mechanism for all polymerases. Nature. 1998;391(6664):231–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

