Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing
- PMID: 20534518
- PMCID: PMC2895082
- DOI: 10.1073/pnas.1006594107
Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing
Abstract
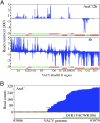
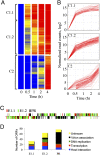
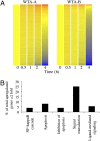
Deep RNA sequencing was used to simultaneously analyze vaccinia virus (VACV) and HeLa cell transcriptomes at progressive times following infection. VACV, the prototypic member of the poxvirus family, replicates in the cytoplasm and contains a double-stranded DNA genome with approximately 200 closely spaced open reading frames (ORFs). The acquisition of a total of nearly 500 million short cDNA sequences allowed construction of temporal strand-specific maps of the entire VACV transcriptome at single-base resolution and analysis of over 14,000 host mRNAs. Before viral DNA replication, transcripts from 118 VACV ORFs were detected; after replication, transcripts from 93 additional ORFs were characterized. The high resolution permitted determination of the precise boundaries of many mRNAs including read-through transcripts and location of mRNA start sites and adjacent promoters. Temporal analysis revealed two clusters of early mRNAs that were synthesized in the presence of inhibitors of protein as well as DNA synthesis, indicating that they do not correspond to separate immediate- and delayed-early classes as defined for other DNA viruses. The proportion of viral RNAs reached 25-55% of the total at 4 h. This rapid change, resulting in a relative decrease of the vast majority of host mRNAs, can contribute to the profound shutdown of host protein synthesis and blunting of antiviral responses. At 2 h, however, a minority of cellular mRNAs was increased. The overrepresented functional categories of the up-regulated RNAs were NF-kappaB cascade, apoptosis, signal transduction, and ligand-mediated signaling, which likely represent the host response to invasion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Deciphering poxvirus gene expression by RNA sequencing and ribosome profiling.J Virol. 2015 Jul;89(13):6874-86. doi: 10.1128/JVI.00528-15. Epub 2015 Apr 22. J Virol. 2015. PMID: 25903347 Free PMC article.
-
Genome-wide analysis of the 5' and 3' ends of vaccinia virus early mRNAs delineates regulatory sequences of annotated and anomalous transcripts.J Virol. 2011 Jun;85(12):5897-909. doi: 10.1128/JVI.00428-11. Epub 2011 Apr 13. J Virol. 2011. PMID: 21490097 Free PMC article.
-
Expression profiling of the intermediate and late stages of poxvirus replication.J Virol. 2011 Oct;85(19):9899-908. doi: 10.1128/JVI.05446-11. Epub 2011 Jul 27. J Virol. 2011. PMID: 21795349 Free PMC article.
-
Organization and expression of the poxvirus genome.Experientia. 1982 Mar 15;38(3):285-97. doi: 10.1007/BF01949349. Experientia. 1982. PMID: 6281055 Review.
-
Virus synthesis and replication: reovirus vs. vaccinia virus.Yale J Biol Med. 1980 Jan-Feb;53(1):27-39. Yale J Biol Med. 1980. PMID: 6990634 Free PMC article. Review.
Cited by
-
A Next-Generation Sequencing Approach Uncovers Viral Transcripts Incorporated in Poxvirus Virions.Viruses. 2017 Oct 13;9(10):296. doi: 10.3390/v9100296. Viruses. 2017. PMID: 29027916 Free PMC article.
-
Construction and Isolation of Recombinant Vaccinia Virus by Homologous Recombination Using Fluorescent Protein Markers.Methods Mol Biol. 2025;2860:83-95. doi: 10.1007/978-1-0716-4160-6_6. Methods Mol Biol. 2025. PMID: 39621262
-
Temporal expression classes and functions of vaccinia virus and mpox (monkeypox) virus genes.mBio. 2025 Apr 9;16(4):e0380924. doi: 10.1128/mbio.03809-24. Epub 2025 Mar 20. mBio. 2025. PMID: 40111027 Free PMC article. Review.
-
Vaccinia Virus Encodes a Novel Inhibitor of Apoptosis That Associates with the Apoptosome.J Virol. 2017 Nov 14;91(23):e01385-17. doi: 10.1128/JVI.01385-17. Print 2017 Dec 1. J Virol. 2017. PMID: 28904196 Free PMC article.
-
Heat-inactivated modified vaccinia virus Ankara boosts Th1 cellular and humoral immunity as a vaccine adjuvant.NPJ Vaccines. 2022 Oct 19;7(1):120. doi: 10.1038/s41541-022-00542-5. NPJ Vaccines. 2022. PMID: 36261460 Free PMC article.
References
-
- Moss B. Poxviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2905–2946.
-
- Davison AJ, Moss B. Structure of vaccinia virus early promoters. J Mol Biol. 1989;210:749–769. - PubMed
-
- Davison AJ, Moss B. Structure of vaccinia virus late promoters. J Mol Biol. 1989;210:771–784. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

