A fluorophore ligase for site-specific protein labeling inside living cells
- PMID: 20534555
- PMCID: PMC2890758
- DOI: 10.1073/pnas.0914067107
A fluorophore ligase for site-specific protein labeling inside living cells
Abstract
Biological microscopy would benefit from smaller alternatives to green fluorescent protein for imaging specific proteins in living cells. Here we introduce PRIME (PRobe Incorporation Mediated by Enzymes), a method for fluorescent labeling of peptide-fused recombinant proteins in living cells with high specificity. PRIME uses an engineered fluorophore ligase, which is derived from the natural Escherichia coli enzyme lipoic acid ligase (LplA). Through structure-guided mutagenesis, we created a mutant ligase capable of recognizing a 7-hydroxycoumarin substrate and catalyzing its covalent conjugation to a transposable 13-amino acid peptide called LAP (LplA Acceptor Peptide). We showed that this fluorophore ligation occurs in cells in 10 min and that it is highly specific for LAP fusion proteins over all endogenous mammalian proteins. By genetically targeting the PRIME ligase to specific subcellular compartments, we were able to selectively label spatially distinct subsets of proteins, such as the surface pool of neurexin and the nuclear pool of actin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

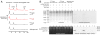

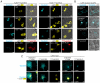
Comment in
-
Protein labeling approaching its PRIME.Nat Methods. 2010 Aug;7(8):584. doi: 10.1038/nmeth0810-584. Nat Methods. 2010. PMID: 20704019 No abstract available.
References
-
- Lin MZ, Wang L. Selective labeling of proteins with chemical probes in living cells. Physiology. 2008;23:131–141. - PubMed
-
- Los GV, et al. HatoTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3:373–382. - PubMed
-
- Gautier A, et al. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15:128–136. - PubMed
-
- Moritz OL, Tam BM, Papermaster DS, Nakayama T. A functional rhodopsin-green fluorescent protein fusion protein localizes correctly in transgenic Xenopus laevis retinal rods and is expressed in a time-dependent pattern. J Biol Chem. 2001;276:28242–28251. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

