Reverse biological engineering of hrdB to enhance the production of avermectins in an industrial strain of Streptomyces avermitilis
- PMID: 20534557
- PMCID: PMC2895102
- DOI: 10.1073/pnas.1006085107
Reverse biological engineering of hrdB to enhance the production of avermectins in an industrial strain of Streptomyces avermitilis
Abstract
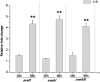
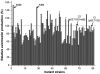
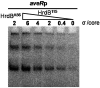
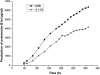
Avermectin and its analogues are produced by the actinomycete Streptomyces avermitilis and are widely used in the field of animal health, agriculture, and human health. Here we have adopted a practical approach to successfully improve avermectin production in an industrial overproducer. Transcriptional levels of the wild-type strain and industrial overproducer in production cultures were monitored using microarray analysis. The avermectin biosynthetic genes, especially the pathway-specific regulatory gene, aveR, were up-regulated in the high-producing strain. The upstream promoter region of aveR was predicted and proved to be directly recognized by sigma(hrdB) in vitro. A mutant library of hrdB gene was constructed by error-prone PCR and selected by high-throughput screening. As a result of evolved hrdB expressed in the modified avermectin high-producing strain, 6.38 g/L of avermectin B1a was produced with over 50% yield improvement, in which the transcription level of aveR was significantly increased. The relevant residues were identified to center in the conserved regions. Engineering of the hrdB gene can not only elicit the overexpression of aveR but also allows for simultaneous transcription of many other genes. The results indicate that manipulating the key genes revealed by reverse engineering can effectively improve the yield of the target metabolites, providing a route to optimize production in these complex regulatory systems.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Inactivation of the extracytoplasmic function sigma factor Sig6 stimulates avermectin production in Streptomyces avermitilis.Biotechnol Lett. 2011 Oct;33(10):1955-61. doi: 10.1007/s10529-011-0673-x. Epub 2011 Jun 21. Biotechnol Lett. 2011. PMID: 21691838
-
Characterization of a regulatory gene, aveR, for the biosynthesis of avermectin in Streptomyces avermitilis.Appl Microbiol Biotechnol. 2009 Apr;82(6):1089-96. doi: 10.1007/s00253-008-1850-2. Epub 2009 Jan 16. Appl Microbiol Biotechnol. 2009. PMID: 19148632
-
Enhanced production of avermectin by deletion of type III polyketide synthases biosynthetic cluster rpp in Streptomyces avermitilis.Lett Appl Microbiol. 2016 Nov;63(5):384-390. doi: 10.1111/lam.12635. Lett Appl Microbiol. 2016. PMID: 27538855
-
Avermectin: biochemical and molecular basis of its biosynthesis and regulation.Appl Microbiol Biotechnol. 2004 Feb;63(6):626-34. doi: 10.1007/s00253-003-1491-4. Epub 2003 Dec 20. Appl Microbiol Biotechnol. 2004. PMID: 14689246 Review.
-
Avermectins and Their Derivatives: Recent Advances in Biosynthesis and Application.J Agric Food Chem. 2025 Jan 22;73(3):1757-1774. doi: 10.1021/acs.jafc.4c07024. Epub 2025 Jan 8. J Agric Food Chem. 2025. PMID: 39772536 Review.
Cited by
-
Recent advances in natural products exploitation in Streptomyces via synthetic biology.Eng Life Sci. 2019 Mar 12;19(6):452-462. doi: 10.1002/elsc.201800137. eCollection 2019 Jun. Eng Life Sci. 2019. PMID: 32625022 Free PMC article. Review.
-
Phosphate limitation increases coenzyme Q10 production in industrial Rhodobacter sphaeroides HY01.Synth Syst Biotechnol. 2019 Nov 30;4(4):212-219. doi: 10.1016/j.synbio.2019.11.001. eCollection 2019 Dec. Synth Syst Biotechnol. 2019. PMID: 31890925 Free PMC article.
-
A novel TetR family transcriptional regulator, SAV576, negatively controls avermectin biosynthesis in Streptomyces avermitilis.PLoS One. 2013 Aug 13;8(8):e71330. doi: 10.1371/journal.pone.0071330. eCollection 2013. PLoS One. 2013. PMID: 23967193 Free PMC article.
-
The Application of Regulatory Cascades in Streptomyces: Yield Enhancement and Metabolite Mining.Front Microbiol. 2020 Mar 24;11:406. doi: 10.3389/fmicb.2020.00406. eCollection 2020. Front Microbiol. 2020. PMID: 32265866 Free PMC article. Review.
-
Verrucisidinol and verrucosidinol acetate, two pyrone-type polyketides isolated from a marine derived fungus, Penicillium aurantiogriseum.Mar Drugs. 2010 Nov 1;8(11):2744-54. doi: 10.3390/md8112744. Mar Drugs. 2010. PMID: 21139842 Free PMC article.
References
-
- Demain AL. Pharmaceutically active secondary metabolites of microorganisms. Appl Microbiol Biotechnol. 1999;52:455–463. - PubMed
-
- Zhang L. In: Natural Products: Drug Discovery and Therapeutic Medicine. Demain AL, Zhang L, editors. Totowa, NJ: Humana Press; 2005. pp. 33–55.
-
- Baltz RH. Genetic methods and strategies for secondary metabolite yield improvement in actinomycetes. Antonie van Leeuwenhoek. 2001;79:251–259. - PubMed
-
- Parekh S, Vinci VA, Strobel RJ. Improvement of microbial strains and fermentation processes. Appl Microbiol Biotechnol. 2000;54:287–301. - PubMed
-
- Rokem JS, Lantz AE, Nielsen J. Systems biology of antibiotic production by microorganisms. Nat Prod Rep. 2007;24:1262–1287. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

