Strong purifying selection at genes escaping X chromosome inactivation
- PMID: 20534706
- PMCID: PMC2981488
- DOI: 10.1093/molbev/msq143
Strong purifying selection at genes escaping X chromosome inactivation
Abstract
To achieve dosage balance of X-linked genes between mammalian males and females, one female X chromosome becomes inactivated. However, approximately 15% of genes on this inactivated chromosome escape X chromosome inactivation (XCI). Here, using a chromosome-wide analysis of primate X-linked orthologs, we test a hypothesis that such genes evolve under a unique selective pressure. We find that escape genes are subject to stronger purifying selection than inactivated genes and that positive selection does not significantly affect the evolution of these genes. The strength of selection does not differ between escape genes with similar versus different expression levels in males versus females. Intriguingly, escape genes possessing Y homologs evolve under the strongest purifying selection. We also found evidence of stronger conservation in gene expression levels in escape than inactivated genes. We hypothesize that divergence in function and expression between X and Y gametologs is driving such strong purifying selection for escape genes.
Figures

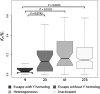
References
-
- Bondy CA. Turner's syndrome and X chromosome-based differences in disease susceptibility. Gend Med. 2006;3:18–30. - PubMed
-
- Burgoyne PS. Mammalian sex determination: thumbs down for zinc finger? Nature. 1989;342:860–862. - PubMed
-
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. - PubMed
-
- Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8:689–698. - PubMed
-
- Ellison JW, Wardak Z, Young MF, Gehron Robey P, Laig-Webster M, Chiong W. PHOG, a candidate gene for involvement in the short stature of Turner syndrome. Hum Mol Genet. 1997;6:1341–1347. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

