The lytic transcriptome of Kaposi's sarcoma-associated herpesvirus reveals extensive transcription of noncoding regions, including regions antisense to important genes
- PMID: 20534856
- PMCID: PMC2916530
- DOI: 10.1128/JVI.00645-10
The lytic transcriptome of Kaposi's sarcoma-associated herpesvirus reveals extensive transcription of noncoding regions, including regions antisense to important genes
Abstract
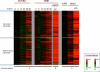
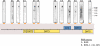
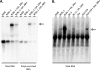
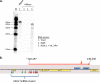
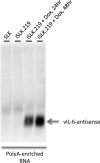
Genomewide analyses of the mammalian transcriptome have revealed that large tracts of sequence previously annotated as noncoding are frequently transcribed and give rise to stable RNA. Although the transcription of individual genes of the Kaposi's sarcoma-associated herpesvirus (KSHV) has been well studied, little is known of the architecture of the viral transcriptome on a genomewide scale. Here we have employed a genomewide tiling array to examine the lytic transcriptome of the Kaposi's sarcoma-associated herpesvirus, KSHV. Our results reveal that during lytic growth (but not during latency), there is extensive transcription from noncoding regions, including both intergenic regions and, especially, noncoding regions antisense to known open reading frames (ORFs). Several of these transcripts have been characterized in more detail, including (i) a 10-kb RNA antisense to the major latency locus, including many of its microRNAs as well as its ORFs; (ii) a 17-kb RNA antisense to numerous ORFs at the left-hand end of the genome; and (iii) a 0.7-kb RNA antisense to the viral homolog of interleukin-6 (vIL-6). These studies indicate that the lytic herpesviral transcriptome resembles a microcosm of the host transcriptome and provides a useful system for the study of noncoding RNAs.
Figures


References
-
- Akusjarvi, G. 2008. Temporal regulation of adenovirus major late alternative RNA splicing. Front. Biosci. 13:5006-5015. - PubMed
-
- Bertone, P., V. Stolc, T. E. Royce, J. S. Rozowsky, A. E. Urban, X. Zhu, J. L. Rinn, W. Tongprasit, M. Samanta, S. Weissman, M. Gerstein, and M. Snyder. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306:2242-2246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

