Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences
- PMID: 20534883
- PMCID: PMC2928500
- DOI: 10.1101/gr.106245.110
Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences
Abstract
The centromere is essential for faithful chromosome segregation by providing the site for kinetochore assembly. Although the role of the centromere is conserved throughout evolution, the DNA sequences associated with centromere regions are highly divergent among species and it remains to be determined how centromere DNA directs kinetochore formation. Despite the active use of chicken DT40 cells in studies of chromosome segregation, the sequence of the chicken centromere was unclear. Here, we performed a comprehensive analysis of chicken centromere DNA which revealed unique features of chicken centromeres compared with previously studied vertebrates. Centromere DNA sequences from the chicken macrochromosomes, with the exception of chromosome 5, contain chromosome-specific homogenous tandem repetitive arrays that span several hundred kilobases. In contrast, the centromeres of chromosomes 5, 27, and Z do not contain tandem repetitive sequences and span non-tandem-repetitive sequences of only approximately 30 kb. To test the function of these centromere sequences, we conditionally removed the centromere from the Z chromosome using genetic engineering and have shown that that the non-tandem-repeat sequence of chromosome Z is a functional centromere.
Figures



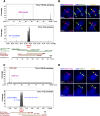

References
-
- Alexandrov I, Kazakov A, Tumeneva I, Shepelev V, Yurov Y 2001. Alpha-satellite DNA of primates: Old and new families. Chromosoma 110: 253–266 - PubMed
-
- Bonner TI, Brenner DJ, Neufield BR, Britten RJ 1973. Reduction in the rate of DNA reassociation by sequence divergence. J Mol Biol 81: 123–135 - PubMed
-
- Carbone L, Nergadze SG, Magnani E, Misceo D, Francesca Cardone M, Roberto R, Bertoni L, Attolini C, Francesca Piras M, de Jong P, et al. 2006. Evolutionary movement of centromeres in horse, donkey, and zebra. Genomics 87: 777–782 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
