Protonation states of the key active site residues and structural dynamics of the glmS riboswitch as revealed by molecular dynamics
- PMID: 20536206
- PMCID: PMC2900856
- DOI: 10.1021/jp9109699
Protonation states of the key active site residues and structural dynamics of the glmS riboswitch as revealed by molecular dynamics
Abstract
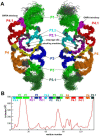
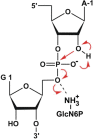
The glmS catalytic riboswitch is part of the 5'-untranslated region of mRNAs encoding glucosamine-6-phosphate (GlcN6P) synthetase (glmS) in numerous gram-positive bacteria. Binding of the cofactor GlcN6P induces site-specific self-cleavage of the RNA. However, the detailed reaction mechanism as well as the protonation state of the glmS reactive form still remains elusive. To probe the dominant protonation states of key active site residues, we carried out explicit solvent molecular dynamic simulations involving various protonation states of three crucial active site moieties observed in the available crystal structures: (i) guanine G40 (following the Thermoanaerobacter tengcongensis numbering), (ii) the GlcN6P amino/ammonium group, and (iii) the GlcN6P phosphate moiety. We found that a deprotonated G40(-) seems incompatible with the observed glmS active site architecture. Our data suggest that the canonical form of G40 plays a structural role by stabilizing an in-line attack conformation of the cleavage site A-1(2'-OH) nucleophile, rather than a more direct chemical role. In addition, we observe weakened cofactor binding upon protonation of the GlcN6P phosphate moiety, which explains the experimentally observed increase in K(m) with decreasing pH. Finally, we discuss a possible role of cofactor binding and its interaction with the G65 and G1 purines in structural stabilization of the A-1(2'-OH) in-line attack conformation. On the basis of the identified dominant protonation state of the reaction precursor, we propose a hypothesis of the self-cleavage mechanism in which A-1(2'-OH) is activated as a nucleophile by the G1(pro-R(p)) nonbridging oxygen of the scissile phosphate, whereas the ammonium group of GlcN6P acts as the general acid protonating the G1(O5') leaving group.
Figures




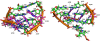


Similar articles
-
Chemical feasibility of the general acid/base mechanism of glmS ribozyme self-cleavage.Biopolymers. 2015 Oct;103(10):550-62. doi: 10.1002/bip.22657. Biopolymers. 2015. PMID: 25858644 Free PMC article.
-
Deciphering the role of glucosamine-6-phosphate in the riboswitch action of glmS ribozyme.RNA. 2010 Dec;16(12):2455-63. doi: 10.1261/rna.2334110. Epub 2010 Oct 22. RNA. 2010. PMID: 20971809 Free PMC article.
-
Structural basis of glmS ribozyme activation by glucosamine-6-phosphate.Science. 2006 Sep 22;313(5794):1752-6. doi: 10.1126/science.1129666. Science. 2006. PMID: 16990543
-
The glmS ribozyme: use of a small molecule coenzyme by a gene-regulatory RNA.Q Rev Biophys. 2010 Nov;43(4):423-47. doi: 10.1017/S0033583510000144. Epub 2010 Sep 8. Q Rev Biophys. 2010. PMID: 20822574 Free PMC article. Review.
-
Catalytic strategies of self-cleaving ribozymes.Acc Chem Res. 2008 Aug;41(8):1027-35. doi: 10.1021/ar800050c. Epub 2008 Jul 25. Acc Chem Res. 2008. PMID: 18652494 Review.
Cited by
-
Towards Accurate Prediction of Protonation Equilibrium of Nucleic Acids.J Phys Chem Lett. 2013 Mar 7;4(5):760-766. doi: 10.1021/jz400078d. Epub 2013 Feb 12. J Phys Chem Lett. 2013. PMID: 23526474 Free PMC article.
-
Molecular dynamics simulations of G-DNA and perspectives on the simulation of nucleic acid structures.Methods. 2012 May;57(1):25-39. doi: 10.1016/j.ymeth.2012.04.005. Epub 2012 Apr 16. Methods. 2012. PMID: 22525788 Free PMC article. Review.
-
Use of a coenzyme by the glmS ribozyme-riboswitch suggests primordial expansion of RNA chemistry by small molecules.Philos Trans R Soc Lond B Biol Sci. 2011 Oct 27;366(1580):2942-8. doi: 10.1098/rstb.2011.0131. Philos Trans R Soc Lond B Biol Sci. 2011. PMID: 21930586 Free PMC article.
-
Chemical feasibility of the general acid/base mechanism of glmS ribozyme self-cleavage.Biopolymers. 2015 Oct;103(10):550-62. doi: 10.1002/bip.22657. Biopolymers. 2015. PMID: 25858644 Free PMC article.
-
Domain motion of individual F1-ATPase β-subunits during unbiased molecular dynamics simulations.J Phys Chem A. 2011 Jun 30;115(25):7267-74. doi: 10.1021/jp2005088. Epub 2011 Apr 1. J Phys Chem A. 2011. PMID: 21452901 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

