Macrocyclic histone deacetylase inhibitors
- PMID: 20536416
- PMCID: PMC3144151
- DOI: 10.2174/156802610792232079
Macrocyclic histone deacetylase inhibitors
Abstract
Histone deacetylase inhibitors (HDACi) are an emerging class of novel anti-cancer drugs that cause growth arrest, differentiation, and apoptosis of tumor cells. In addition, they have shown promise as anti-parasitic, anti-neurodegenerative, anti-rheumatologic and immunosuppressant agents. To date, several structurally distinct small molecule HDACi have been reported including aryl hydroxamates, benzamides, short-chain fatty acids, electrophilic ketones, and macrocyclic peptides. Macrocyclic HDACi possess the most complex cap-groups which interact with HDAC enzyme's outer rim and have demonstrated excellent HDAC inhibition potency and isoform selectivity. This review focuses on the recent progress and current state of macrocyclic HDACi.
Figures


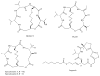
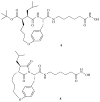
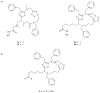





Similar articles
-
Recent progress in the development of histone deacetylase inhibitors as anti-cancer agents.Mini Rev Med Chem. 2013 Dec;13(14):1999-2013. doi: 10.2174/13895575113136660102. Mini Rev Med Chem. 2013. PMID: 24160707 Review.
-
Non-peptide macrocyclic histone deacetylase inhibitors.J Med Chem. 2009 Jan 22;52(2):456-68. doi: 10.1021/jm801128g. J Med Chem. 2009. PMID: 19093884 Free PMC article.
-
Non-peptide macrocyclic histone deacetylase inhibitors derived from tricyclic ketolide skeleton.J Med Chem. 2010 Aug 26;53(16):6100-11. doi: 10.1021/jm100507q. J Med Chem. 2010. PMID: 20669972 Free PMC article.
-
Natural and Synthetic Macrocyclic Inhibitors of the Histone Deacetylase Enzymes.Chembiochem. 2017 Jan 3;18(1):5-49. doi: 10.1002/cbic.201600519. Epub 2016 Dec 8. Chembiochem. 2017. PMID: 27748555 Review.
-
Structure of 'linkerless' hydroxamic acid inhibitor-HDAC8 complex confirms the formation of an isoform-specific subpocket.J Struct Biol. 2016 Sep;195(3):373-378. doi: 10.1016/j.jsb.2016.06.023. Epub 2016 Jun 29. J Struct Biol. 2016. PMID: 27374062 Free PMC article.
Cited by
-
Synthesis of HDAC Substrate Peptidomimetic Inhibitors Using Fmoc Amino Acids Incorporating Zinc-Binding Groups.Org Lett. 2019 May 3;21(9):3178-3182. doi: 10.1021/acs.orglett.9b00885. Epub 2019 Apr 18. Org Lett. 2019. PMID: 30998366 Free PMC article.
-
Vorinostat suppresses hypoxia signaling by modulating nuclear translocation of hypoxia inducible factor 1 alpha.Oncotarget. 2017 May 23;8(34):56110-56125. doi: 10.18632/oncotarget.18125. eCollection 2017 Aug 22. Oncotarget. 2017. PMID: 28915577 Free PMC article.
-
Latency Reversing Agents: Kick and Kill of HTLV-1?Int J Mol Sci. 2021 May 24;22(11):5545. doi: 10.3390/ijms22115545. Int J Mol Sci. 2021. PMID: 34073995 Free PMC article. Review.
-
A Review on the Therapeutic Role of TKIs in Case of CML in Combination With Epigenetic Drugs.Front Genet. 2021 Oct 22;12:742802. doi: 10.3389/fgene.2021.742802. eCollection 2021. Front Genet. 2021. PMID: 34745216 Free PMC article. Review.
-
FDA-approved drugs featuring macrocycles or medium-sized rings.Arch Pharm (Weinheim). 2025 Jan;358(1):e2400890. doi: 10.1002/ardp.202400890. Arch Pharm (Weinheim). 2025. PMID: 39865335 Free PMC article. Review.
References
-
- Carey N, La Thangue NB. Histone deacetyalse inhibitors: Gathering pace. Curr Opin Pharmacol. 2006;6:369–375. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
-
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. - PubMed
-
- Althaus FR, Höfferer L, Kleczkowska HE, Malanga M, Naegeli H, Panzeter PL, Realini CA. Histone Shuttling by Poly ADP-Ribosylation. Mol Cell Biochem. 1994;138:53–59. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

