Granzyme A activates another way to die
- PMID: 20536557
- PMCID: PMC2905780
- DOI: 10.1111/j.0105-2896.2010.00902.x
Granzyme A activates another way to die
Abstract
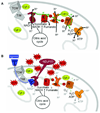
Granzyme A (GzmA) is the most abundant serine protease in killer cell cytotoxic granules. GzmA activates a novel programed cell death pathway that begins in the mitochondrion, where cleavage of NDUFS3 in electron transport complex I disrupts mitochondrial metabolism and generates reactive oxygen species (ROS). ROS drives the endoplasmic reticulum-associated SET complex into the nucleus, where it activates single-stranded DNA damage. GzmA also targets other important nuclear proteins for degradation, including histones, the lamins that maintain the nuclear envelope, and several key DNA damage repair proteins (Ku70, PARP-1). Cells that are resistant to the caspases or GzmB by overexpressing bcl-2 family anti-apoptotic proteins or caspase or GzmB protease inhibitors are sensitive to GzmA. By activating multiple cell death pathways, killer cells provide better protection against a variety of intracellular pathogens and tumors. GzmA also has proinflammatory activity; it activates pro-interleukin-1beta and may also have other proinflammatory effects that remain to be elucidated.
Figures



Similar articles
-
Granzyme a, a stealth killer in the mitochondrion.Cell. 2008 May 16;133(4):568-70. doi: 10.1016/j.cell.2008.04.031. Cell. 2008. PMID: 18485862
-
Granzyme A cleaves a mitochondrial complex I protein to initiate caspase-independent cell death.Cell. 2008 May 16;133(4):681-92. doi: 10.1016/j.cell.2008.03.032. Cell. 2008. PMID: 18485875 Free PMC article.
-
Orphan granzymes find a home.Immunol Rev. 2010 May;235(1):117-27. doi: 10.1111/j.0105-2896.2010.00889.x. Immunol Rev. 2010. PMID: 20536559 Review.
-
Granzymes A and B directly cleave lamins and disrupt the nuclear lamina during granule-mediated cytolysis.Proc Natl Acad Sci U S A. 2001 May 8;98(10):5746-51. doi: 10.1073/pnas.101329598. Epub 2001 May 1. Proc Natl Acad Sci U S A. 2001. PMID: 11331782 Free PMC article.
-
Nuclear war: the granzyme A-bomb.Curr Opin Immunol. 2003 Oct;15(5):553-9. doi: 10.1016/s0952-7915(03)00108-0. Curr Opin Immunol. 2003. PMID: 14499264 Review.
Cited by
-
Deep profiling and custom databases improve detection of proteoforms generated by alternative splicing.Genome Res. 2019 Dec;29(12):2046-2055. doi: 10.1101/gr.248435.119. Epub 2019 Nov 14. Genome Res. 2019. PMID: 31727681 Free PMC article.
-
Granzyme A Stimulates pDCs to Promote Adaptive Immunity via Induction of Type I IFN.Front Immunol. 2019 Jun 26;10:1450. doi: 10.3389/fimmu.2019.01450. eCollection 2019. Front Immunol. 2019. PMID: 31293597 Free PMC article.
-
CRISPR/Cas13d targeting GZMA in PARs pathway regulates the function of osteoclasts in chronic apical periodontitis.Cell Mol Biol Lett. 2023 Aug 25;28(1):70. doi: 10.1186/s11658-023-00477-2. Cell Mol Biol Lett. 2023. PMID: 37626297 Free PMC article.
-
Serine protease inhibition attenuates rIL-12-induced GZMA activity and proinflammatory events by modulating the Th2 profile from estrogen-treated mice.Endocrinology. 2014 Aug;155(8):2909-23. doi: 10.1210/en.2014-1045. Epub 2014 May 19. Endocrinology. 2014. PMID: 24840346 Free PMC article.
-
Plasma proteomic profiles of UK Biobank participants with multiple sclerosis.Ann Clin Transl Neurol. 2024 Mar;11(3):698-709. doi: 10.1002/acn3.51990. Epub 2024 Jan 28. Ann Clin Transl Neurol. 2024. PMID: 38282238 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

