TIM genes: a family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity
- PMID: 20536563
- PMCID: PMC2914464
- DOI: 10.1111/j.0105-2896.2010.00903.x
TIM genes: a family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity
Abstract
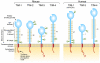
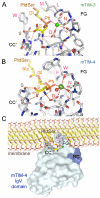
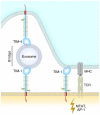
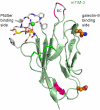
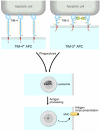
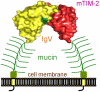
The TIM (T cell/transmembrane, immunoglobulin, and mucin) gene family plays a critical role in regulating immune responses, including allergy, asthma, transplant tolerance, autoimmunity, and the response to viral infections. The unique structure of TIM immunoglobulin variable region domains allows highly specific recognition of phosphatidylserine (PtdSer), exposed on the surface of apoptotic cells. TIM-1, TIM-3, and TIM-4 all recognize PtdSer but differ in expression, suggesting that they have distinct functions in regulating immune responses. TIM-1, an important susceptibility gene for asthma and allergy, is preferentially expressed on T-helper 2 (Th2) cells and functions as a potent costimulatory molecule for T-cell activation. TIM-3 is preferentially expressed on Th1 and Tc1 cells, and generates an inhibitory signal resulting in apoptosis of Th1 and Tc1 cells. TIM-3 is also expressed on some dendritic cells and can mediate phagocytosis of apoptotic cells and cross-presentation of antigen. In contrast, TIM-4 is exclusively expressed on antigen-presenting cells, where it mediates phagocytosis of apoptotic cells and plays an important role in maintaining tolerance. TIM molecules thus provide a functional repertoire for recognition of apoptotic cells, which determines whether apoptotic cell recognition leads to immune activation or tolerance, depending on the TIM molecule engaged and the cell type on which it is expressed.
Figures
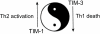


References
-
- McIntire JJ, et al. Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim gene family. Nat Immunol. 2001;2:1109–1116. - PubMed
-
- McIntire JJ, Umetsu DT, DeKruyff RH. TIM-1, a novel allergy and asthma susceptibility gene. Springer Semin Immunopathol. 2004;25:335–348. - PubMed
-
- Marsh DG, et al. Linkage analysis of IL4 and other chromosome 5q31.1 markers and total serum immunoglobulin E concentrations. Science. 1994;264:1152–1156. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

