mu-CS: an extension of the TM4 platform to manage Affymetrix binary data
- PMID: 20537149
- PMCID: PMC2907348
- DOI: 10.1186/1471-2105-11-315
mu-CS: an extension of the TM4 platform to manage Affymetrix binary data
Abstract
Background: A main goal in understanding cell mechanisms is to explain the relationship among genes and related molecular processes through the combined use of technological platforms and bioinformatics analysis. High throughput platforms, such as microarrays, enable the investigation of the whole genome in a single experiment. There exist different kind of microarray platforms, that produce different types of binary data (images and raw data). Moreover, also considering a single vendor, different chips are available. The analysis of microarray data requires an initial preprocessing phase (i.e. normalization and summarization) of raw data that makes them suitable for use on existing platforms, such as the TIGR M4 Suite. Nevertheless, the annotations of data with additional information such as gene function, is needed to perform more powerful analysis. Raw data preprocessing and annotation is often performed in a manual and error prone way. Moreover, many available preprocessing tools do not support annotation. Thus novel, platform independent, and possibly open source tools enabling the semi-automatic preprocessing and annotation of microarray data are needed.
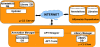
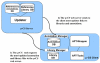
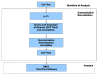
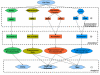
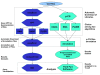
Results: The paper presents mu-CS (Microarray Cel file Summarizer), a cross-platform tool for the automatic normalization, summarization and annotation of Affymetrix binary data. mu-CS is based on a client-server architecture. The mu-CS client is provided both as a plug-in of the TIGR M4 platform and as a Java standalone tool and enables users to read, preprocess and analyse binary microarray data, avoiding the manual invocation of external tools (e.g. the Affymetrix Power Tools), the manual loading of preprocessing libraries, and the management of intermediate files. The mu-CS server automatically updates the references to the summarization and annotation libraries that are provided to the mu-CS client before the preprocessing. The mu-CS server is based on the web services technology and can be easily extended to support more microarray vendors (e.g. Illumina).
Conclusions: Thus mu-CS users can directly manage binary data without worrying about locating and invoking the proper preprocessing tools and chip-specific libraries. Moreover, users of the mu-CS plugin for TM4 can manage Affymetrix binary files without using external tools, such as APT (Affymetrix Power Tools) and related libraries. Consequently, mu-CS offers four main advantages: (i) it avoids to waste time for searching the correct libraries, (ii) it reduces possible errors in the preprocessing and further analysis phases, e.g. due to the incorrect choice of parameters or the use of old libraries, (iii) it implements the annotation of preprocessed data, and finally, (iv) it may enhance the quality of further analysis since it provides the most updated annotation libraries. The mu-CS client is freely available as a plugin of the TM4 platform as well as a standalone application at the project web site (http://bioingegneria.unicz.it/M-CS).
Figures



References
-
- Affymetrix website. http://www.affymetrix.com
-
- Durinck S. Pre-processing of microarray data and analysis of differential expression. Methods in molecular biology (Clifton, N.J.) 2008;452:89–110. full_text. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

