The ubiquitin-conjugating enzyme HR6B is required for maintenance of X chromosome silencing in mouse spermatocytes and spermatids
- PMID: 20537150
- PMCID: PMC3091626
- DOI: 10.1186/1471-2164-11-367
The ubiquitin-conjugating enzyme HR6B is required for maintenance of X chromosome silencing in mouse spermatocytes and spermatids
Abstract
Background: The ubiquitin-conjugating enzyme HR6B is required for spermatogenesis in mouse. Loss of HR6B results in aberrant histone modification patterns on the trancriptionally silenced X and Y chromosomes (XY body) and on centromeric chromatin in meiotic prophase. We studied the relationship between these chromatin modifications and their effects on global gene expression patterns, in spermatocytes and spermatids.
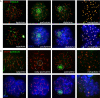
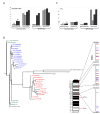
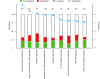
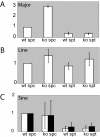
Results: HR6B is enriched on the XY body and on centromeric regions in pachytene spermatocytes. Global gene expression analyses revealed that spermatid-specific single- and multicopy X-linked genes are prematurely expressed in Hr6b knockout spermatocytes. Very few other differences in gene expression were observed in these cells, except for upregulation of major satellite repeat transcription. In contrast, in Hr6b knockout spermatids, 7298 genes were differentially expressed; 65% of these genes was downregulated, but we observed a global upregulation of gene transcription from the X chromosome. In wild type spermatids, approximately 20% of the single-copy X-linked genes reach an average expression level that is similar to the average expression from autosomes.
Conclusions: Spermatids maintain an enrichment of repressive chromatin marks on the X chromosome, originating from meiotic prophase, but this does not interfere with transcription of the single-copy X-linked genes that are reactivated or specifically activated in spermatids. HR6B represses major satellite repeat transcription in spermatocytes, and functions in the maintenance of X chromosome silencing in spermatocytes and spermatids. It is discussed that these functions involve modification of chromatin structure, possibly including H2B ubiquitylation.
Figures

Similar articles
-
Increased phosphorylation and dimethylation of XY body histones in the Hr6b-knockout mouse is associated with derepression of the X chromosome.J Cell Sci. 2007 Jun 1;120(Pt 11):1841-51. doi: 10.1242/jcs.03451. Epub 2007 May 8. J Cell Sci. 2007. PMID: 17488778
-
UBR2 mediates transcriptional silencing during spermatogenesis via histone ubiquitination.Proc Natl Acad Sci U S A. 2010 Feb 2;107(5):1912-7. doi: 10.1073/pnas.0910267107. Epub 2010 Jan 11. Proc Natl Acad Sci U S A. 2010. PMID: 20080676 Free PMC article.
-
Histone ubiquitination and chromatin remodeling in mouse spermatogenesis.Dev Biol. 1999 Mar 15;207(2):322-33. doi: 10.1006/dbio.1998.9155. Dev Biol. 1999. PMID: 10068466
-
Histone crotonylation specifically marks the haploid male germ cell gene expression program: post-meiotic male-specific gene expression.Bioessays. 2012 Mar;34(3):187-93. doi: 10.1002/bies.201100141. Epub 2011 Dec 15. Bioessays. 2012. PMID: 22170506 Review.
-
The role of chromatin modifications in progression through mouse meiotic prophase.J Genet Genomics. 2014 Mar 20;41(3):97-106. doi: 10.1016/j.jgg.2014.01.003. Epub 2014 Jan 22. J Genet Genomics. 2014. PMID: 24656230 Review.
Cited by
-
Role of the ubiquitin-proteasome system in nervous system function and disease: using C. elegans as a dissecting tool.Cell Mol Life Sci. 2012 Aug;69(16):2691-715. doi: 10.1007/s00018-012-0946-0. Epub 2012 Mar 3. Cell Mol Life Sci. 2012. PMID: 22382927 Free PMC article. Review.
-
SPIN1 is a proto-oncogene and SPIN3 is a tumor suppressor in human seminoma.Oncotarget. 2018 Aug 21;9(65):32466-32477. doi: 10.18632/oncotarget.25977. eCollection 2018 Aug 21. Oncotarget. 2018. PMID: 30197756 Free PMC article.
-
UBE2B mRNA alterations are associated with severe oligozoospermia in infertile men.Mol Hum Reprod. 2013 Jun;19(6):388-94. doi: 10.1093/molehr/gat008. Epub 2013 Jan 31. Mol Hum Reprod. 2013. PMID: 23378580 Free PMC article.
-
Round Spermatid Injection Rescues Female Lethality of a Paternally Inherited Xist Deletion in Mouse.PLoS Genet. 2016 Oct 7;12(10):e1006358. doi: 10.1371/journal.pgen.1006358. eCollection 2016 Oct. PLoS Genet. 2016. PMID: 27716834 Free PMC article.
-
Destruction or Reconstruction: A Subtle Liaison between the Proteolytic and Signaling Role of Protein Ubiquitination in Spermatogenesis.Adv Exp Med Biol. 2021;1288:215-240. doi: 10.1007/978-3-030-77779-1_11. Adv Exp Med Biol. 2021. PMID: 34453739
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

