Time-lapse imaging and cell-specific expression profiling reveal dynamic branching and molecular determinants of a multi-dendritic nociceptor in C. elegans
- PMID: 20537990
- PMCID: PMC2919608
- DOI: 10.1016/j.ydbio.2010.05.502
Time-lapse imaging and cell-specific expression profiling reveal dynamic branching and molecular determinants of a multi-dendritic nociceptor in C. elegans
Abstract
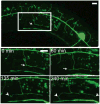
Nociceptive neurons innervate the skin with complex dendritic arbors that respond to pain-evoking stimuli such as harsh mechanical force or extreme temperatures. Here we describe the structure and development of a model nociceptor, the PVD neuron of C. elegans, and identify transcription factors that control morphogenesis of the PVD dendritic arbor. The two PVD neuron cell bodies occupy positions on either the right (PVDR) or left (PVDL) sides of the animal in posterior-lateral locations. Imaging with a GFP reporter revealed a single axon projecting from the PVD soma to the ventral cord and an elaborate, highly branched arbor of dendritic processes that envelop the animal with a web-like array directly beneath the skin. Dendritic branches emerge in a step-wise fashion during larval development and may use an existing network of peripheral nerve cords as guideposts for key branching decisions. Time-lapse imaging revealed that branching is highly dynamic with active extension and withdrawal and that PVD branch overlap is prevented by a contact-dependent self-avoidance, a mechanism that is also employed by sensory neurons in other organisms. With the goal of identifying genes that regulate dendritic morphogenesis, we used the mRNA-tagging method to produce a gene expression profile of PVD during late larval development. This microarray experiment identified>2,000 genes that are 1.5X elevated relative to all larval cells. The enriched transcripts encode a wide range of proteins with potential roles in PVD function (e.g., DEG/ENaC and Trp channels) or development (e.g., UNC-5 and LIN-17/frizzled receptors). We used RNAi and genetic tests to screen 86 transcription factors from this list and identified eleven genes that specify PVD dendritic structure. These transcription factors appear to control discrete steps in PVD morphogenesis and may either promote or limit PVD branching at specific developmental stages. For example, time-lapse imaging revealed that MEC-3 (LIM homeodomain) is required for branch initiation in early larval development whereas EGL-44 (TEAD domain) prevents ectopic PVD branching in the adult. A comparison of PVD-enriched transcripts to a microarray profile of mammalian nociceptors revealed homologous genes with potentially shared nociceptive functions. We conclude that PVD neurons display striking structural, functional and molecular similarities to nociceptive neurons from more complex organisms and can thus provide a useful model system in which to identify evolutionarily conserved determinants of nociceptor fate.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures









Similar articles
-
Mechanisms that regulate morphogenesis of a highly branched neuron in C. elegans.Dev Biol. 2019 Jul 1;451(1):53-67. doi: 10.1016/j.ydbio.2019.04.002. Epub 2019 Apr 17. Dev Biol. 2019. PMID: 31004567 Free PMC article. Review.
-
Separate transcriptionally regulated pathways specify distinct classes of sister dendrites in a nociceptive neuron.Dev Biol. 2017 Dec 15;432(2):248-257. doi: 10.1016/j.ydbio.2017.10.009. Epub 2017 Oct 13. Dev Biol. 2017. PMID: 29031632 Free PMC article.
-
C. elegans bicd-1, homolog of the Drosophila dynein accessory factor Bicaudal D, regulates the branching of PVD sensory neuron dendrites.Development. 2011 Feb;138(3):507-18. doi: 10.1242/dev.060939. Development. 2011. PMID: 21205795 Free PMC article.
-
The proprotein convertase KPC-1/furin controls branching and self-avoidance of sensory dendrites in Caenorhabditis elegans.PLoS Genet. 2014 Sep 18;10(9):e1004657. doi: 10.1371/journal.pgen.1004657. eCollection 2014 Sep. PLoS Genet. 2014. PMID: 25232734 Free PMC article.
-
Dendrite morphogenesis in Caenorhabditis elegans.Genetics. 2024 Jun 5;227(2):iyae056. doi: 10.1093/genetics/iyae056. Genetics. 2024. PMID: 38785371 Free PMC article. Review.
Cited by
-
High-Dose Ionizing Radiation Impairs Healthy Dendrite Growth in C. elegans.Adv Radiat Oncol. 2023 Dec 5;9(3):101415. doi: 10.1016/j.adro.2023.101415. eCollection 2024 Mar. Adv Radiat Oncol. 2023. PMID: 38379892 Free PMC article.
-
MBL-1/Muscleblind regulates neuronal differentiation and controls the splicing of a terminal selector in Caenorhabditis elegans.PLoS Genet. 2024 Oct 18;20(10):e1011276. doi: 10.1371/journal.pgen.1011276. eCollection 2024 Oct. PLoS Genet. 2024. PMID: 39423233 Free PMC article.
-
Identification of healthspan-promoting genes in Caenorhabditis elegans based on a human GWAS study.Biogerontology. 2022 Aug;23(4):431-452. doi: 10.1007/s10522-022-09969-8. Epub 2022 Jun 24. Biogerontology. 2022. PMID: 35748965 Free PMC article.
-
Mechanisms that regulate morphogenesis of a highly branched neuron in C. elegans.Dev Biol. 2019 Jul 1;451(1):53-67. doi: 10.1016/j.ydbio.2019.04.002. Epub 2019 Apr 17. Dev Biol. 2019. PMID: 31004567 Free PMC article. Review.
-
Sequence determinants of the Caenhorhabditis elegans dopamine transporter dictating in vivo axonal export and synaptic localization.Mol Cell Neurosci. 2017 Jan;78:41-51. doi: 10.1016/j.mcn.2016.11.011. Epub 2016 Nov 30. Mol Cell Neurosci. 2017. PMID: 27913309 Free PMC article.
References
-
- Abrahamsen B, Zhao J, Asante CO, Cendan CM, Marsh S, Martinez-Barbera JP, Nassar MA, Dickenson AH, Wood JN. The cell and molecular basis of mechanical, cold, and inflammatory pain. Science. 2008;321:702–705. - PubMed
-
- Baker MW, Macagno ER. In vivo imaging of growth cone and filopodial dynamics: evidence for contact-mediated retraction of filopodia leading to the tiling of sibling processes. J Comp Neurol. 2007;500:850–862. - PubMed
-
- Barr MM, DeModena J, Braun D, Nguyen CQ, Hall DH, Sternberg PW. The Caenorhabditis elegans autosomal dominant polycystic kidney disease gene homologs lov-1 and pkd-2 act in the same pathway. Curr Biol. 2001;11:1341–1346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 DK058404/DK/NIDDK NIH HHS/United States
- P30 HD015052/HD/NICHD NIH HHS/United States
- P30 EY008126/EY/NEI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- F31 NS49743/NS/NINDS NIH HHS/United States
- P60 DK20593/DK/NIDDK NIH HHS/United States
- HD15052/HD/NICHD NIH HHS/United States
- P30 DK58404/DK/NIDDK NIH HHS/United States
- P30EY08126/EY/NEI NIH HHS/United States
- P60 DK020593/DK/NIDDK NIH HHS/United States
- R21 NS066882/NS/NINDS NIH HHS/United States
- F31 NS049743/NS/NINDS NIH HHS/United States
- R01 NS026115/NS/NINDS NIH HHS/United States
- P30 CA68485/CA/NCI NIH HHS/United States
- R21 NS06882/NS/NINDS NIH HHS/United States
- R01 NS26115/NS/NINDS NIH HHS/United States
- P01 HL6744/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

