Pgrmc1 (progesterone receptor membrane component 1) associates with epidermal growth factor receptor and regulates erlotinib sensitivity
- PMID: 20538600
- PMCID: PMC2915713
- DOI: 10.1074/jbc.M110.134585
Pgrmc1 (progesterone receptor membrane component 1) associates with epidermal growth factor receptor and regulates erlotinib sensitivity
Abstract
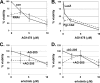
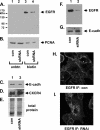
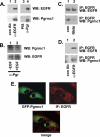
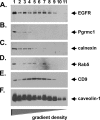
Tumorigenesis requires the concerted action of multiple pathways, including pathways that stimulate proliferation and metabolism. Epidermal growth factor receptor (EGFR) is a transmembrane receptor-tyrosine kinase that is associated with cancer progression, and the EGFR inhibitors erlotinib/tarceva and tyrphostin/AG-1478 are potent anti-cancer therapeutics. Pgrmc1 (progesterone receptor membrane component 1) is a cytochrome b(5)-related protein that is up-regulated in tumors and promotes cancer growth. Pgrmc1 and its homologues have been implicated in cell signaling, and we show here that Pgrmc1 increases susceptibility to AG-1478 and erlotinib, increases plasma membrane EGFR levels, and co-precipitates with EGFR. Pgrmc1 co-localizes with EGFR in cytoplasmic vesicles and co-fractionates with EGFR in high density microsomes. The findings have therapeutic potential because a Pgrmc1 small molecule ligand, which inhibits growth in a variety of cancer cell types, de-stabilized EGFR in multiple tumor cell lines. EGFR is one of the most potent receptor-tyrosine kinases driving tumorigenesis, and our data support a role for Pgrmc1 in promoting several cancer phenotypes at least in part by binding EGFR and stabilizing plasma membrane pools of the receptor.
Figures


Similar articles
-
Progesterone receptor membrane component 1 leads to erlotinib resistance, initiating crosstalk of Wnt/β-catenin and NF-κB pathways, in lung adenocarcinoma cells.Sci Rep. 2020 Mar 16;10(1):4748. doi: 10.1038/s41598-020-61727-3. Sci Rep. 2020. PMID: 32179851 Free PMC article.
-
Neutrophil gelatinase-associated lipocalin (NGAL) expression is dependent on the tumor-associated sigma-2 receptor S2RPgrmc1.J Biol Chem. 2012 Apr 27;287(18):14494-501. doi: 10.1074/jbc.M111.324921. Epub 2012 Mar 14. J Biol Chem. 2012. PMID: 22418433 Free PMC article.
-
Rapamycin synergizes with the epidermal growth factor receptor inhibitor erlotinib in non-small-cell lung, pancreatic, colon, and breast tumors.Mol Cancer Ther. 2006 Nov;5(11):2676-84. doi: 10.1158/1535-7163.MCT-06-0166. Mol Cancer Ther. 2006. PMID: 17121914
-
S2R(Pgrmc1): the cytochrome-related sigma-2 receptor that regulates lipid and drug metabolism and hormone signaling.Expert Opin Drug Metab Toxicol. 2012 Mar;8(3):361-70. doi: 10.1517/17425255.2012.658367. Epub 2012 Feb 1. Expert Opin Drug Metab Toxicol. 2012. PMID: 22292588 Review.
-
PGRMC1 (progesterone receptor membrane component 1): a targetable protein with multiple functions in steroid signaling, P450 activation and drug binding.Pharmacol Ther. 2009 Jan;121(1):14-9. doi: 10.1016/j.pharmthera.2008.09.006. Epub 2008 Nov 1. Pharmacol Ther. 2009. PMID: 18992768 Free PMC article. Review.
Cited by
-
Plasminogen activator inhibitor 1 RNA-binding protein interacts with progesterone receptor membrane component 1 to regulate progesterone's ability to maintain the viability of spontaneously immortalized granulosa cells and rat granulosa cells.Biol Reprod. 2013 Jan 25;88(1):20. doi: 10.1095/biolreprod.112.103036. Print 2013 Jan. Biol Reprod. 2013. PMID: 23242527 Free PMC article.
-
Progesterone receptor membrane component 1 (PGRMC1) binds and stabilizes cytochromes P450 through a heme-independent mechanism.J Biol Chem. 2021 Nov;297(5):101316. doi: 10.1016/j.jbc.2021.101316. Epub 2021 Oct 20. J Biol Chem. 2021. PMID: 34678314 Free PMC article.
-
Progesterone Receptor Membrane Component 1 suppresses the p53 and Wnt/β-catenin pathways to promote human pluripotent stem cell self-renewal.Sci Rep. 2018 Feb 14;8(1):3048. doi: 10.1038/s41598-018-21322-z. Sci Rep. 2018. PMID: 29445107 Free PMC article.
-
Progesterone receptor membrane component 1 promotes survival of human breast cancer cells and the growth of xenograft tumors.Cancer Biol Ther. 2016;17(3):262-71. doi: 10.1080/15384047.2016.1139240. Epub 2016 Jan 19. Cancer Biol Ther. 2016. PMID: 26785864 Free PMC article.
-
Progesterone receptor membrane component 1 is phosphorylated upon progestin treatment in breast cancer cells.Oncotarget. 2017 Aug 2;8(42):72480-72493. doi: 10.18632/oncotarget.19819. eCollection 2017 Sep 22. Oncotarget. 2017. PMID: 29069804 Free PMC article.
References
-
- Ritter C. A., Arteaga C. L. (2003) Semin. Oncol. 30, 3–11 - PubMed
-
- Salomon D. S., Brandt R., Ciardiello F., Normanno N. (1995) Crit. Rev. Oncol. Hematol. 19, 183–232 - PubMed
-
- Ono M., Kuwano M. (2006) Clin. Cancer Res. 12, 7242–7251 - PubMed
-
- Bareschino M. A., Schettino C., Troiani T., Martinelli E., Morgillo F., Ciardiello F. (2007) Ann. Oncol. 18, vi35–41 - PubMed
-
- Lynch T. J., Bell D. W., Sordella R., Gurubhagavatula S., Okimoto R. A., Brannigan B. W., Harris P. L., Haserlat S. M., Supko J. G., Haluska F. G., Louis D. N., Christiani D. C., Settleman J., Haber D. A. (2004) N. Engl. J. Med. 350, 2129–2139 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

