Cyclin regulation by the s phase checkpoint
- PMID: 20538605
- PMCID: PMC2924074
- DOI: 10.1074/jbc.M110.138669
Cyclin regulation by the s phase checkpoint
Abstract
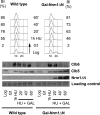
In eukaryotic cells a surveillance mechanism, the S phase checkpoint, detects and responds to DNA damage and replication stress, protecting DNA replication and arresting cell cycle progression. We show here that the S phase cyclins Clb5 and Clb6 are regulated in response to genotoxic stress in the budding yeast Saccharomyces cerevisiae. Clb5 and Clb6 are responsible for the activation of the specific Cdc28 cyclin-dependent kinase activity that drives the onset and progression of the S phase. Intriguingly, Clb5 and Clb6 are regulated by different mechanisms. Thus, the presence of Clb6, which is eliminated early in an unperturbed S phase, is stabilized when replication is compromised by replication stress or DNA damage. Such stabilization depends on the checkpoint kinases Mec1 and Rad53. The stabilization of Clb6 levels is a dynamic process that requires continued de novo protein synthesis, because the cyclin remains subject to degradation. It also requires the activity of the G(1) transcription factor Mlu1 cell cycle box-binding factor (MBF) in the S phase, whereas Dun1, the checkpoint kinase characteristically responsible for the transcriptional response to genotoxic stress, is dispensable in this case. On the other hand, two subpopulations of endogenous Clb5 can be distinguished according to turnover in an unperturbed S phase. In the presence of replication stress, the unstable Clb5 pool is stabilized, and such stabilization requires neither MBF transcriptional activity nor de novo protein synthesis.
Figures


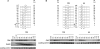
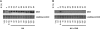






Similar articles
-
Cdk1-interacting protein Cip1 is regulated by the S phase checkpoint in response to genotoxic stress.Genes Cells. 2017 Oct;22(10):850-860. doi: 10.1111/gtc.12518. Epub 2017 Aug 3. Genes Cells. 2017. PMID: 28771906 Free PMC article.
-
S-phase cyclin-dependent kinases promote sister chromatid cohesion in budding yeast.Mol Cell Biol. 2011 Jun;31(12):2470-83. doi: 10.1128/MCB.05323-11. Epub 2011 Apr 25. Mol Cell Biol. 2011. PMID: 21518961 Free PMC article.
-
CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint.Genes Dev. 1998 Sep 1;12(17):2698-710. doi: 10.1101/gad.12.17.2698. Genes Dev. 1998. PMID: 9732268 Free PMC article.
-
The S-phase checkpoint: targeting the replication fork.Biol Cell. 2009 Aug 19;101(11):617-27. doi: 10.1042/BC20090053. Biol Cell. 2009. PMID: 19686094 Review.
-
Mechanisms involved in regulating DNA replication origins during the cell cycle and in response to DNA damage.Philos Trans R Soc Lond B Biol Sci. 2004 Jan 29;359(1441):31-8. doi: 10.1098/rstb.2003.1362. Philos Trans R Soc Lond B Biol Sci. 2004. PMID: 15065654 Free PMC article. Review.
Cited by
-
The DNA damage checkpoint response to replication stress: A Game of Forks.Front Genet. 2013 Mar 13;4:26. doi: 10.3389/fgene.2013.00026. eCollection 2013. Front Genet. 2013. PMID: 23493417 Free PMC article.
-
Research progress of ECT2 and RhoA-related signaling pathways in gynecological tumors.Front Cell Dev Biol. 2025 Jun 27;13:1602649. doi: 10.3389/fcell.2025.1602649. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40655948 Free PMC article. Review.
-
G1 cyclin driven DNA replication.Cell Cycle. 2015;14(24):3842-50. doi: 10.1080/15384101.2015.1070995. Cell Cycle. 2015. PMID: 26176277 Free PMC article.
-
Cdk1-interacting protein Cip1 is regulated by the S phase checkpoint in response to genotoxic stress.Genes Cells. 2017 Oct;22(10):850-860. doi: 10.1111/gtc.12518. Epub 2017 Aug 3. Genes Cells. 2017. PMID: 28771906 Free PMC article.
-
Regulation of CLB6 expression by the cytoplasmic deadenylase Ccr4 through its coding and 3' UTR regions.PLoS One. 2022 May 6;17(5):e0268283. doi: 10.1371/journal.pone.0268283. eCollection 2022. PLoS One. 2022. PMID: 35522675 Free PMC article.
References
-
- Hartwell L. H., Weinert T. A. (1989) Science 246, 629–634 - PubMed
-
- Weinert T. A., Hartwell L. H. (1988) Science 241, 317–322 - PubMed
-
- Hartwell L. H., Kastan M. B. (1994) Science 266, 1821–1828 - PubMed
-
- Bartkova J., Horejsí Z., Koed K., Krämer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J. M., Lukas C., Ørntoft T., Lukas J., Bartek J. (2005) Nature 434, 864–870 - PubMed
-
- Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L. V., Kolettas E., Niforou K., Zoumpourlis V. C., Takaoka M., Nakagawa H., Tort F., Fugger K., Johansson F., Sehested M., Andersen C. L., Dyrskjot L., Ørntoft T., Lukas J., Kittas C., Helleday T., Halazonetis T. D., Bartek J., Gorgoulis V. G. (2006) Nature 444, 633–637 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

