In planta gene expression analysis of Xanthomonas oryzae pathovar oryzae, African strain MAI1
- PMID: 20540733
- PMCID: PMC2893596
- DOI: 10.1186/1471-2180-10-170
In planta gene expression analysis of Xanthomonas oryzae pathovar oryzae, African strain MAI1
Abstract
Background: Bacterial leaf blight causes significant yield losses in rice crops throughout Asia and Africa. Although both the Asian and African strains of the pathogen, Xanthomonas oryzae pv. oryzae (Xoo), induce similar symptoms, they are nevertheless genetically different, with the African strains being more closely related to the Asian X. oryzae pv. oryzicola (Xoc).
Results: Changes in gene expression of the African Xoo strain MAI1 in the susceptible rice cultivar Nipponbare were profiled, using an SSH Xoo DNA microarray. Microarray hybridization was performed comparing bacteria recovered from plant tissues at 1, 3, and 6 days after inoculation (dai) with bacteria grown in vitro. A total of 710 bacterial genes were found to be differentially expressed, with 407 up-regulated and 303 down-regulated. Expression profiling indicated that less than 20% of the 710 bacterial transcripts were induced in the first 24 h after inoculation, whereas 63% were differentially expressed at 6 dai. The 710 differentially expressed genes were one-end sequenced. 535 sequences were obtained from which 147 non-redundant sequences were identified. Differentially expressed genes were related to metabolism, secretion and transport, pathogen adherence to plant tissues, plant cell-wall degradation, IS elements, and virulence. In addition, various other genes encoding proteins with unknown function or showing no similarity to other proteins were also induced. The Xoo MAI1 non-redundant set of sequences was compared against several X. oryzae genomes, revealing a specific group of genes that was present only in MAI1. Numerous IS elements were also found to be differentially expressed. Quantitative real-time PCR confirmed 86% of the identified profile on a set of 14 genes selected according to the microarray analysis.
Conclusions: This is the first report to compare the expression of Xoo genes in planta across different time points during infection. This work shows that as-yet-unidentified and potentially new virulence factors are appearing in an emerging African pathogen. It also confirms that African Xoo strains do differ from their Asian counterparts, even at the transcriptional level.
Figures
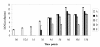


References
-
- Séré Y, Onasanya A, Verdier V, Akator K, Ouédraogo L, Segda Z, Coulibaly M, Sido A, Basso A. Rice Bacterial Leaf Blight in West Africa: Preliminary Studies on Disease in Farmers' Fields and Screening Released Varieties for Resistance to the Bacteria. Asian Journal of Plant Sciences. 2005;4:577–579. doi: 10.3923/ajps.2005.577.579. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

