High-throughput production of human proteins for crystallization: the SGC experience
- PMID: 20541610
- PMCID: PMC2938586
- DOI: 10.1016/j.jsb.2010.06.008
High-throughput production of human proteins for crystallization: the SGC experience
Abstract
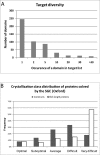
Producing purified human proteins with high yield and purity remains a considerable challenge. We describe the methods utilized in the Structural Genomics Consortium (SGC) in Oxford, resulting in successful purification of 48% of human proteins attempted; of those, the structures of approximately 40% were solved by X-ray crystallography. The main driver has been the parallel processing of multiple (typically 9-20) truncated constructs of each target; modest diversity in vectors and host systems; and standardized purification procedures. We provide method details as well as data on the properties of the constructs leading to crystallized proteins and the impact of methodological variants. These can be used to formulate guidelines for initial approaches to expression of new eukaryotic proteins.
Figures





References
-
- Abbott A. Protein structures hint at the shape of things to come. Nature. 2005;435:547. - PubMed
-
- Banci L., Bertini I., Cusack S., de Jong R.N., Heinemann U., Jones E.Y., Kozielski F., Maskos K., Messerschmidt A., Owens R., Perrakis A., Poterszman A., Schneider G., Siebold C., Silman I., Sixma T., Stewart-Jones G., Sussman J.L., Thierry J.C., Moras D. First steps towards effective methods in exploiting high-throughput technologies for the determination of human protein structures of high biomedical value. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1208–1217. - PubMed
-
- Berrow N.S., Bussow K., Coutard B., Diprose J., Ekberg M., Folkers G.E., Levy N., Lieu V., Owens R.J., Peleg Y., Pinaglia C., Quevillon-Cheruel S., Salim L., Scheich C., Vincentelli R., Busso D. Recombinant protein expression and solubility screening in Escherichia coli: a comparative study. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1218–1226. - PubMed
-
- Burgess-Brown N.A., Sharma S., Sobott F., Loenarz C., Oppermann U., Gileadi O. Codon optimization can improve expression of human genes in Escherichia coli: a multi-gene study. Protein Expr. Purif. 2008;59:94–102. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

