The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly
- PMID: 20542003
- PMCID: PMC3372891
- DOI: 10.1016/j.molcel.2010.05.024
The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly
Abstract
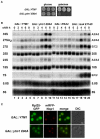
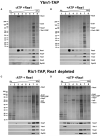
The AAA(+)-ATPase Rea1 removes the ribosome biogenesis factor Rsa4 from pre-60S ribosomal subunits in the nucleoplasm to drive nuclear export of the subunit. To do this, Rea1 utilizes a MIDAS domain to bind a conserved motif in Rsa4. Here, we show that the Rea1 MIDAS domain binds another pre-60S factor, Ytm1, via a related motif. In vivo Rea1 contacts Ytm1 before it contacts Rsa4, and its interaction with Ytm1 coincides with the exit of early pre-60S particles from the nucleolus to the nucleoplasm. In vitro, Rea1's ATPase activity triggers removal of the conserved nucleolar Ytm1-Erb1-Nop7 subcomplex from isolated early pre-60S particle. We suggest that the Rea1 AAA(+)-ATPase functions at successive maturation steps to remove ribosomal factors at critical transition points, first driving the exit of early pre-60S particles from the nucleolus and then driving late pre-60S particles from the nucleus.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures




References
-
- Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. - PubMed
-
- Bassler J, Kallas M, Hurt E. The NUG1 GTPase reveals and N-terminal RNA-binding domain that is essential for association with 60 S pre-ribosomal particles. J. Biol. Chem. 2006;281:24737–24744. - PubMed
-
- Davies JM, Brunger AT, Weis WI. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–726. - PubMed
-
- DeLaBarre B, Brunger AT. Nucleotide dependent motion and mechanism of action of p97/VCP. J. Mol. Biol. 2005;347:437–452. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

