Cost function masking during normalization of brains with focal lesions: still a necessity?
- PMID: 20542122
- PMCID: PMC2938189
- DOI: 10.1016/j.neuroimage.2010.06.003
Cost function masking during normalization of brains with focal lesions: still a necessity?
Abstract
Although normalization of brain images is critical to the analysis of structural damage across individuals, loss of tissue due to focal lesions presents challenges to the available normalization algorithms. Until recently, cost function masking, as advocated by Brett and colleagues (2001), was the accepted method to overcome difficulties encountered when normalizing damaged brains; however, development of the unified segmentation approach for normalization in SPM5 (Ashburner & Friston, 2005) offered an alternative. Crinion et al. (2007) demonstrated this approach produced normalization results without cost function masking that appeared to be robust to lesion effects when tested using the same simulated lesions studied by Brett et al. (2001). The present study sought to confirm the validity of this approach in brains with focal damage due to vascular events. To do so, we examined outcomes of normalization using unified segmentation with and without cost function masking in 49 brain images with chronic stroke. Lesion masks were created using two approaches (precise and rough drawings of lesion boundaries), and normalization was implemented with both smoothed and unsmoothed versions of the masks. We found that failure to employ cost function masking produced less accurate results in real and simulated lesions, compared to masked normalization, both in terms of deformation field displacement and voxelwise intensity differences. Additionally, unmasked normalization led to significant underestimation of lesion volume relative to all four masking conditions, especially in patients with large lesions. Taken together, these findings suggest cost function masking is still necessary when normalizing brain images with chronic infarcts.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
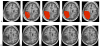




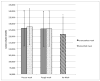
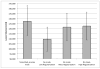
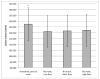
References
-
- Ashburner J, Friston KJ. Unified segmentation. NeuroImage. 2005;26:839–851. - PubMed
-
- Brett M, Leff AP, Rorden C, Ashburner J. Spatial normalization of brain images with focal lesions using cost function masking. NeuroImage. 2001;14(2):486–500. - PubMed
-
- Kim J, Avants B, Patel S, Whyte J. Spatial normalization of injured brains for neuroimaging research: An illustrative introduction of available options. 2007. http://www.ncrrn.org/papers/methodology_papers/sp_norm_kim.pdf.
-
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang M-C, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage. 2009;46:786–802. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

