Binding site number variation and high-affinity binding consensus of Myb-SANT-like transcription factor Adf-1 in Drosophilidae
- PMID: 20542916
- PMCID: PMC2965233
- DOI: 10.1093/nar/gkq504
Binding site number variation and high-affinity binding consensus of Myb-SANT-like transcription factor Adf-1 in Drosophilidae
Abstract
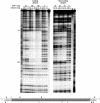
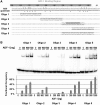
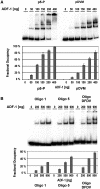
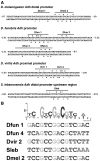
There is a growing interest in the evolution of transcription factor binding sites and corresponding functional change of transcriptional regulation. In this context, we have examined the structural changes of the ADF-1 binding sites at the Adh promoters of Drosophila funebris and D. virilis. We detected an expanded footprinted region in D. funebris that contains various adjacent binding sites with different binding affinities. ADF-1 was described to direct sequence-specific DNA binding to sites consisting of the multiple trinucleotide repeat . The ADF-1 recognition sites with high binding affinity differ from this trinucleotide repeat consensus sequence and a new consensus sequence is proposed for the high-affinity ADF-1 binding sites. In vitro transcription experiments with the D. funebris and D. virilis ADF-1 binding regions revealed that stronger ADF-1 binding to the expanded D. funebris ADF-1 binding region only moderately lead to increased transcriptional activity of the Adh gene. The potential of this regional expansion is discussed in the context of different ADF-1 cellular concentrations and maintenance of the ADF-1 stimulus. Altogether, evolutionary change of ADF-1 binding regions involves both, rearrangements of complex binding site cluster and also nucleotide substitutions within sites that lead to different binding affinities.
Figures

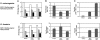

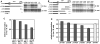
References
-
- King M-C, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. - PubMed
-
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. - PubMed
-
- Ludwig MZ. Functional evolution of noncoding DNA. Curr. Opin. Genet. Dev. 2002;12:634–639. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

