Hydroxyapatite-mediated separation of double-stranded DNA, single-stranded DNA, and RNA genomes from natural viral assemblages
- PMID: 20543058
- PMCID: PMC2916501
- DOI: 10.1128/AEM.00204-10
Hydroxyapatite-mediated separation of double-stranded DNA, single-stranded DNA, and RNA genomes from natural viral assemblages
Abstract
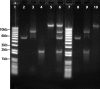
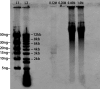
Metagenomics can be used to determine the diversity of complex, often unculturable, viral communities with various nucleic acid compositions. Here, we report the use of hydroxyapatite chromatography to efficiently fractionate double-stranded DNA (dsDNA), single-stranded DNA (ssDNA), dsRNA, and ssRNA genomes from known bacteriophages. Linker-amplified shotgun libraries were constructed to generate sequencing reads from each hydroxyapatite fraction. Greater than 90% of the reads displayed significant similarity to the expected genomes at the nucleotide level. These methods were applied to marine viruses collected from the Chesapeake Bay and the Dry Tortugas National Park. Isolated nucleic acids were fractionated using hydroxyapatite chromatography followed by linker-amplified shotgun library construction and sequencing. Taxonomic analysis demonstrated that the majority of environmental sequences, regardless of their source nucleic acid, were most similar to dsDNA viruses, reflecting the bias of viral metagenomic sequence databases.
Figures


References
-
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne, P. G. Amanatides, S. E. Scherer, P. W. Li, R. A. Hoskins, R. F. Galle, R. A. George, S. E. Lewis, S. Richards, M. Ashburner, S. N. Henderson, G. G. Sutton, J. R. Wortman, M. D. Yandell, Q. Zhang, L. X. Chen, R. C. Brandon, Y. H. Rogers, R. G. Blazej, M. Champe, B. D. Pfeiffer, K. H. Wan, C. Doyle, E. G. Baxter, G. Helt, C. R. Nelson, G. L. Gabor, J. F. Abril, A. Agbayani, H. J. An, C. Andrews-Pfannkoch, D. Baldwin, R. M. Ballew, A. Basu, J. Baxendale, L. Bayraktaroglu, E. M. Beasley, K. Y. Beeson, P. V. Benos, B. P. Berman, D. Bhandari, S. Bolshakov, D. Borkova, M. R. Botchan, J. Bouck, P. Brokstein, P. Brottier, K. C. Burtis, D. A. Busam, H. Butler, E. Cadieu, A. Center, I. Chandra, J. M. Cherry, S. Cawley, C. Dahlke, L. B. Davenport, P. Davies, B. de Pablos, A. Delcher, Z. Deng, A. D. Mays, I. Dew, S. M. Dietz, K. Dodson, L. E. Doup, M. Downes, S. Dugan-Rocha, B. C. Dunkov, P. Dunn, K. J. Durbin, C. C. Evangelista, C. Ferraz, S. Ferriera, W. Fleischmann, C. Fosler, A. E. Gabrielian, N. S. Garg, W. M. Gelbart, K. Glasser, A. Glodek, F. Gong, J. H. Gorrell, Z. Gu, P. Guan, M. Harris, N. L. Harris, D. Harvey, T. J. Heiman, J. R. Hernandez, J. Houck, D. Hostin, K. A. Houston, T. J. Howland, M. H. Wei, C. Ibegwam, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. - PubMed
-
- Angly, F. E., B. Felts, M. Breitbart, P. Salamon, R. A. Edwards, C. Carlson, A. M. Chan, M. Haynes, S. Kelley, H. Liu, J. M. Mahaffy, J. E. Mueller, J. Nulton, R. Olson, R. Parsons, S. Rayhawk, C. A. Suttle, and F. Rohwer. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. - PMC - PubMed
-
- Badger, J. H., T. R. Hoover, Y. V. Brun, R. M. Weiner, M. T. Laub, G. Alexandre, J. Mrazek, Q. Ren, I. T. Paulsen, K. E. Nelson, H. M. Khouri, D. Radune, J. Sosa, R. J. Dodson, S. A. Sullivan, M. J. Rosovitz, R. Madupu, L. M. Brinkac, A. S. Durkin, S. C. Daugherty, S. P. Kothari, M. G. Giglio, L. Zhou, D. H. Haft, J. D. Selengut, T. M. Davidsen, Q. Yang, N. Zafar, and N. L. Ward. 2006. Comparative genomic evidence for a close relationship between the dimorphic prosthecate bacteria Hyphomonas neptunium and Caulobacter crescentus. J. Bacteriol. 188:6841-6850. - PMC - PubMed
-
- Bernardi, G. 1965. Chromatography of nucleic acids on hydroxyapatite. Nature 206:779-783. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

