beta(1-3)Glucanosyltransferase Gel4p is essential for Aspergillus fumigatus
- PMID: 20543062
- PMCID: PMC2918925
- DOI: 10.1128/EC.00107-10
beta(1-3)Glucanosyltransferase Gel4p is essential for Aspergillus fumigatus
Abstract
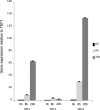
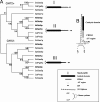
The beta(1-3)glucanosyltransferase GEL family of Aspergillus fumigatus contains 7 genes, among which only 3 are expressed during mycelial growth. The role of the GEL4 gene was investigated in this study. Like the other Gelps, it encodes a glycosylphosphatidylinositol (GPI)-anchored protein. In contrast to the other beta(1-3)glucanosyltransferases analyzed to date, it is essential for this fungal species.
Figures

Similar articles
-
The Aspergillus fumigatus β-1,3-glucanosyltransferase Gel7 plays a compensatory role in maintaining cell wall integrity under stress conditions.Glycobiology. 2014 May;24(5):418-27. doi: 10.1093/glycob/cwu003. Epub 2014 Jan 14. Glycobiology. 2014. PMID: 24429506
-
Deletion of GEL2 encoding for a beta(1-3)glucanosyltransferase affects morphogenesis and virulence in Aspergillus fumigatus.Mol Microbiol. 2005 Jun;56(6):1675-88. doi: 10.1111/j.1365-2958.2005.04654.x. Mol Microbiol. 2005. PMID: 15916615
-
Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall.J Biol Chem. 2000 May 19;275(20):14882-9. doi: 10.1074/jbc.275.20.14882. J Biol Chem. 2000. PMID: 10809732
-
GPI Anchored Proteins in Aspergillus fumigatus and Cell Wall Morphogenesis.Curr Top Microbiol Immunol. 2020;425:167-186. doi: 10.1007/82_2020_207. Curr Top Microbiol Immunol. 2020. PMID: 32418035 Review.
-
Specific molecular features in the organization and biosynthesis of the cell wall of Aspergillus fumigatus.Med Mycol. 2005 May;43 Suppl 1:S15-22. doi: 10.1080/13693780400029155. Med Mycol. 2005. PMID: 16110787 Review.
Cited by
-
Transcriptome and biochemical analysis reveals that suppression of GPI-anchor synthesis leads to autophagy and possible necroptosis in Aspergillus fumigatus.PLoS One. 2013;8(3):e59013. doi: 10.1371/journal.pone.0059013. Epub 2013 Mar 18. PLoS One. 2013. PMID: 23527074 Free PMC article.
-
Polysome profiling reveals broad translatome remodeling during endoplasmic reticulum (ER) stress in the pathogenic fungus Aspergillus fumigatus.BMC Genomics. 2014 Feb 25;15:159. doi: 10.1186/1471-2164-15-159. BMC Genomics. 2014. PMID: 24568630 Free PMC article.
-
Endoplasmic reticulum localized PerA is required for cell wall integrity, azole drug resistance, and virulence in Aspergillus fumigatus.Mol Microbiol. 2014 Jun;92(6):1279-98. doi: 10.1111/mmi.12626. Epub 2014 May 9. Mol Microbiol. 2014. PMID: 24779420 Free PMC article.
-
Redox signalling from NADPH oxidase targets metabolic enzymes and developmental proteins in Fusarium graminearum.Mol Plant Pathol. 2019 Jan;20(1):92-106. doi: 10.1111/mpp.12742. Epub 2018 Nov 7. Mol Plant Pathol. 2019. PMID: 30113774 Free PMC article.
-
The Glycosylphosphatidylinositol-Anchored DFG Family Is Essential for the Insertion of Galactomannan into the β-(1,3)-Glucan-Chitin Core of the Cell Wall of Aspergillus fumigatus.mSphere. 2019 Jul 31;4(4):e00397-19. doi: 10.1128/mSphere.00397-19. mSphere. 2019. PMID: 31366710 Free PMC article.
References
-
- Barral P., Suarez C., Batanero E., Alfonso C., Alché J. D., Rodríguez-Garcia M. I., Villalba M., Rivas G., Rodríguez R. 2005. An olive pollen protein with allergenic activity, Ole e 10, defines a novel family of carbohydrate-binding modules and is potentially implicated in pollen germination. Biochem. J. 390:77–84 - PMC - PubMed
-
- Beauvais A., Drake R., Ng K., Diaquin M., Latgé J. P. 1993. Characterization of the 1,3 β glucan synthase of A. fumigatus. J. Gen. Microbiol. 139:3071–3078 - PubMed
-
- da Silva Ferreira M. E., Kress M. R. V. Z., Savoldi M., Goldman M. H. S., Hartl A., Heinekamp T., Brakhage A. A., Goldman G. H. 2006. The akuBKU80 mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5:207–211 - PMC - PubMed
-
- de Medina-Redondo M., Arnàiz-Pita Y., Fontaine T., Del Rey F., Latge J. P., de Aldana C. R. 2008. The beta-1,3-glucanosyltransferase gas4p is essential for ascospore wall maturation and spore viability in Schizosaccharomyces pombe. Mol. Microbiol. 68:1283–1299 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

