Phosphorylation and dephosphorylation among Dif chemosensory proteins essential for exopolysaccharide regulation in Myxococcus xanthus
- PMID: 20543066
- PMCID: PMC2937368
- DOI: 10.1128/JB.00403-10
Phosphorylation and dephosphorylation among Dif chemosensory proteins essential for exopolysaccharide regulation in Myxococcus xanthus
Abstract
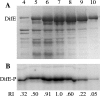
Myxococcus xanthus social gliding motility, which is powered by type IV pili, requires the presence of exopolysaccharides (EPS) on the cell surface. The Dif chemosensory system is essential for the regulation of EPS production. It was demonstrated previously that DifA (methyl-accepting chemotaxis protein [MCP]-like), DifC (CheW-like), and DifE (CheA-like) stimulate whereas DifD (CheY-like) and DifG (CheC-like) inhibit EPS production. DifD was found not to function downstream of DifE in EPS regulation, as a difD difE double mutant phenocopied the difE single mutant. It has been proposed that DifA, DifC, and DifE form a ternary signaling complex that positively regulates EPS production through the kinase activity of DifE. DifD was proposed as a phosphate sink of phosphorylated DifE (DifE approximately P), while DifG would augment the function of DifD as a phosphatase of phosphorylated DifD (DifD approximately P). Here we report in vitro phosphorylation studies with all the Dif chemosensory proteins that were expressed and purified from Escherichia coli. DifE was demonstrated to be an autokinase. Consistent with the formation of a DifA-DifC-DifE complex, DifA and DifC together, but not individually, were found to influence DifE autophosphorylation. DifD, which did not inhibit DifE autophosphorylation directly, was found to accept phosphate from autophosphorylated DifE. While DifD approximately P has an unusually long half-life for dephosphorylation in vitro, DifG efficiently dephosphorylated DifD approximately P as a phosphatase. These results support a model where DifE complexes with DifA and DifC to regulate EPS production through phosphorylation of a downstream target, while DifD and DifG function synergistically to divert phosphates away from DifE approximately P.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

