Discovery and Characterization of HemQ: an essential heme biosynthetic pathway component
- PMID: 20543190
- PMCID: PMC2923992
- DOI: 10.1074/jbc.M110.142604
Discovery and Characterization of HemQ: an essential heme biosynthetic pathway component
Abstract
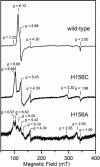
Here we identify a previously undescribed protein, HemQ, that is required for heme synthesis in Gram-positive bacteria. We have characterized HemQ from Bacillus subtilis and a number of Actinobacteria. HemQ is a multimeric heme-binding protein. Spectroscopic studies indicate that this heme is high spin ferric iron and is ligated by a conserved histidine with the sixth coordination site available for binding a small molecule. The presence of HemQ along with the terminal two pathway enzymes, protoporphyrinogen oxidase (HemY) and ferrochelatase, is required to synthesize heme in vivo and in vitro. Although the exact role played by HemQ remains to be characterized, to be fully functional in vitro it requires the presence of a bound heme. HemQ possesses minimal peroxidase activity, but as a catalase it has a turnover of over 10(4) min(-1). We propose that this activity may be required to eliminate hydrogen peroxide that is generated by each turnover of HemY. Given the essential nature of heme synthesis and the restricted distribution of HemQ, this protein is a potential antimicrobial target for pathogens such as Mycobacterium tuberculosis.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

