The C-terminal alpha-alpha superhelix of Pat is required for mRNA decapping in metazoa
- PMID: 20543818
- PMCID: PMC2910274
- DOI: 10.1038/emboj.2010.124
The C-terminal alpha-alpha superhelix of Pat is required for mRNA decapping in metazoa
Abstract
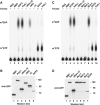
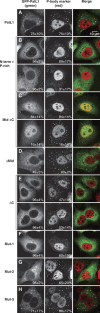
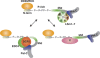
Pat proteins regulate the transition of mRNAs from a state that is translationally active to one that is repressed, committing targeted mRNAs to degradation. Pat proteins contain a conserved N-terminal sequence, a proline-rich region, a Mid domain and a C-terminal domain (Pat-C). We show that Pat-C is essential for the interaction with mRNA decapping factors (i.e. DCP2, EDC4 and LSm1-7), whereas the P-rich region and Mid domain have distinct functions in modulating these interactions. DCP2 and EDC4 binding is enhanced by the P-rich region and does not require LSm1-7. LSm1-7 binding is assisted by the Mid domain and is reduced by the P-rich region. Structural analysis revealed that Pat-C folds into an alpha-alpha superhelix, exposing conserved and basic residues on one side of the domain. This conserved and basic surface is required for RNA, DCP2, EDC4 and LSm1-7 binding. The multiplicity of interactions mediated by Pat-C suggests that certain of these interactions are mutually exclusive and, therefore, that Pat proteins switch decapping partners allowing transitions between sequential steps in the mRNA decapping pathway.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

