Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription
- PMID: 20543824
- PMCID: PMC2892551
- DOI: 10.1038/nature09095
Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription
Abstract
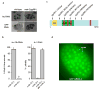
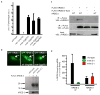
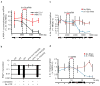
Eukaryotic cells express a wide variety of endogenous small regulatory RNAs that regulate heterochromatin formation, developmental timing, defence against parasitic nucleic acids and genome rearrangement. Many small regulatory RNAs are thought to function in nuclei. For instance, in plants and fungi, short interfering RNA (siRNAs) associate with nascent transcripts and direct chromatin and/or DNA modifications. To understand further the biological roles of small regulatory RNAs, we conducted a genetic screen to identify factors required for RNA interference (RNAi) in Caenorhabditis elegans nuclei. Here we show that the gene nuclear RNAi defective-2 (nrde-2) encodes an evolutionarily conserved protein that is required for siRNA-mediated silencing in nuclei. NRDE-2 associates with the Argonaute protein NRDE-3 within nuclei and is recruited by NRDE-3/siRNA complexes to nascent transcripts that have been targeted by RNAi. We find that nuclear-localized siRNAs direct an NRDE-2-dependent silencing of pre-messenger RNAs (pre-mRNAs) 3' to sites of RNAi, an NRDE-2-dependent accumulation of RNA polymerase (RNAP) II at genomic loci targeted by RNAi, and NRDE-2-dependent decreases in RNAP II occupancy and RNAP II transcriptional activity 3' to sites of RNAi. These results define NRDE-2 as a component of the nuclear RNAi machinery and demonstrate that metazoan siRNAs can silence nuclear-localized RNAs co-transcriptionally. In addition, these results establish a novel mode of RNAP II regulation: siRNA-directed recruitment of NRDE factors that inhibit RNAP II during the elongation phase of transcription.
Figures

Comment in
-
RNA silencing: Nuclear RNAi in worms.Nat Rev Mol Cell Biol. 2010 Aug;11(8):539. doi: 10.1038/nrm2939. Epub 2010 Jun 30. Nat Rev Mol Cell Biol. 2010. PMID: 20588295 No abstract available.
Similar articles
-
An Argonaute transports siRNAs from the cytoplasm to the nucleus.Science. 2008 Jul 25;321(5888):537-41. doi: 10.1126/science.1157647. Science. 2008. PMID: 18653886 Free PMC article.
-
A Conserved NRDE-2/MTR-4 Complex Mediates Nuclear RNAi in Caenorhabditis elegans.Genetics. 2020 Dec;216(4):1071-1085. doi: 10.1534/genetics.120.303631. Epub 2020 Oct 14. Genetics. 2020. PMID: 33055090 Free PMC article.
-
A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation.PLoS Genet. 2011 Aug;7(8):e1002249. doi: 10.1371/journal.pgen.1002249. Epub 2011 Aug 25. PLoS Genet. 2011. PMID: 21901112 Free PMC article.
-
A new layer of rRNA regulation by small interference RNAs and the nuclear RNAi pathway.RNA Biol. 2017 Nov 2;14(11):1492-1498. doi: 10.1080/15476286.2017.1341034. Epub 2017 Jul 21. RNA Biol. 2017. PMID: 28640690 Free PMC article. Review.
-
Biology and Mechanisms of Short RNAs in Caenorhabditis elegans.Adv Genet. 2013;83:1-69. doi: 10.1016/B978-0-12-407675-4.00001-8. Adv Genet. 2013. PMID: 23890211 Review.
Cited by
-
Complex coding of endogenous siRNA, transcriptional silencing and H3K9 methylation on native targets of germline nuclear RNAi in C. elegans.BMC Genomics. 2014 Dec 22;15(1):1157. doi: 10.1186/1471-2164-15-1157. BMC Genomics. 2014. PMID: 25534009 Free PMC article.
-
Argonaute proteins couple chromatin silencing to alternative splicing.Nat Struct Mol Biol. 2012 Oct;19(10):998-1004. doi: 10.1038/nsmb.2373. Epub 2012 Sep 9. Nat Struct Mol Biol. 2012. PMID: 22961379
-
RSR-2, the Caenorhabditis elegans ortholog of human spliceosomal component SRm300/SRRM2, regulates development by influencing the transcriptional machinery.PLoS Genet. 2013 Jun;9(6):e1003543. doi: 10.1371/journal.pgen.1003543. Epub 2013 Jun 6. PLoS Genet. 2013. PMID: 23754964 Free PMC article.
-
A long noncoding RNA positively regulates CD56 in human natural killer cells.Oncotarget. 2016 Nov 8;7(45):72546-72558. doi: 10.18632/oncotarget.12466. Oncotarget. 2016. PMID: 27713137 Free PMC article.
-
Dual roles for nuclear RNAi Argonautes in Caenorhabditis elegans dosage compensation.Genetics. 2022 May 5;221(1):iyac033. doi: 10.1093/genetics/iyac033. Genetics. 2022. PMID: 35234908 Free PMC article.
References
-
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6(1):24–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

