Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity
- PMID: 20543849
- PMCID: PMC2893244
- DOI: 10.1038/ng.605
Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity
Abstract
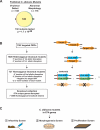
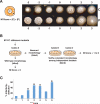
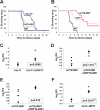
Candida albicans is the most common cause of serious fungal disease in humans. Creation of isogenic null mutants of this diploid organism, which requires sequential gene targeting, allows dissection of virulence mechanisms. Published analyses of such mutants show a near-perfect correlation between C. albicans pathogenicity and the ability to undergo a yeast-to-hypha morphological switch in vitro. However, most studies have used mutants constructed with a marker that is itself a virulence determinant and therefore complicates their interpretation. Using alternative markers, we created approximately 3,000 homozygous deletion strains affecting 674 genes, or roughly 11% of the C. albicans genome. Screening for infectivity in a mouse model and for morphological switching and cell proliferation in vitro, we identified 115 infectivity-attenuated mutants, of which nearly half demonstrated normal morphological switching and proliferation. Analysis of such mutants revealed that virulence requires the glycolipid glucosylceramide. To our knowledge, this is the first C. albicans small molecule that has been found to be required specifically for virulence.
Figures



References
-
- Edmond MB, et al. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis. 1999;29:239–44. - PubMed
-
- Zaoutis TE, et al. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin Infect Dis. 2005;41:1232–9. - PubMed
-
- Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

