Fatty acid amide hydrolase as a potential therapeutic target for the treatment of pain and CNS disorders
- PMID: 20544003
- PMCID: PMC2882713
- DOI: 10.1517/17460440903018857
Fatty acid amide hydrolase as a potential therapeutic target for the treatment of pain and CNS disorders
Abstract
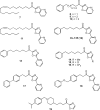
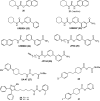
BACKGROUND: Fatty acid amide hydrolase (FAAH) is an integral membrane enzyme that hydrolyzes the endocannabinoid anandamide and related amidated signaling lipids. Genetic or pharmacological inactivation of FAAH produces analgesic, anti-inflammatory, anxiolytic, and antidepressant phenotypes without showing the undesirable side effects of direct cannabinoid receptor agonists, indicating that FAAH may be a promising therapeutic target. OBJECTIVES: This review highlights advances in the development of FAAH inhibitors of different mechanistic classes and their in vivo efficacy. Also highlighted are advances in technology for the in vitro and in vivo selectivity assessment of FAAH inhibitors employing activity-based protein profiling (ABPP) and click chemistry-ABPP, respectively. Recent reports on structure-based drug design for human FAAH generated by protein engineering using interspecies active site conversion are also discussed. METHODS: The literature searches of Medline and SciFinder databases were used. CONCLUSIONS: There has been tremendous progress in our understanding in FAAH and development of FAAH inhibitors with in vivo efficacy, selectivity, and drug like pharmacokinetic properties.
Figures
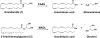



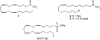




References
-
- Di Marzo V, Bisogno T, De Petrocellis L. Endocannabinoids and related compounds: walking back and forth between plant natural products and animal physiology. Chem Biol. 2007 Jul;14(7):741–56. - PubMed
-
- Fowler CJ. The cannabinoid system and its pharmacological manipulation--a review, with emphasis upon the uptake and hydrolysis of anandamide. Fundam Clin Pharmacol. 2006 Dec;20(6):549–62. - PubMed
-
- Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996 Nov 7;384(6604):83–7. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
