The regulation of class IA PI 3-kinases by inter-subunit interactions
- PMID: 20544340
- PMCID: PMC3234103
- DOI: 10.1007/82_2010_52
The regulation of class IA PI 3-kinases by inter-subunit interactions
Abstract
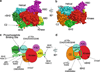
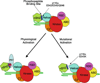
Phosphoinositide 3-kinases (PI 3-kinases) are activated by growth factor and hormone receptors, and regulate cell growth, survival, motility, and responses to changes in nutritional conditions (Engelman et al. 2006). PI 3-kinases have been classified according to their subunit composition and their substrate specificity for phosphoinositides (Vanhaesebroeck et al. 2001). The class IA PI 3-kinase is a heterodimer consisting of one regulatory subunit (p85α, p85β, p55α, p50α, or p55γ) and one 110-kDa catalytic subunit (p110α, β or δ). The Class IB PI 3-kinase is also a dimer, composed of one regulatory subunit (p101 or p87) and one catalytic subunit (p110γ) (Wymann et al. 2003). Class I enzymes will utilize PI, PI[4]P, or PI[4,5]P2 as substrates in vitro, but are thought to primarily produce PI[3,4,5]P3 in cells.The crystal structure of the Class IB PI 3-kinase catalytic subunit p110γ was solved in 1999 (Walker et al. 1999), and crystal or NMR structures of the Class IA p110α catalytic subunit and all of the individual domains of the Class IA p85α regulatory subunit have been solved (Booker et al. 1992; Günther et al. 1996; Hoedemaeker et al. 1999; Huang et al. 2007; Koyama et al. 1993; Miled et al. 2007; Musacchio et al. 1996; Nolte et al. 1996; Siegal et al. 1998). However, a structure of an intact PI 3-kinase enzyme has remained elusive. In spite of this, studies over the past 10 years have lead to important insights into how the enzyme is regulated under physiological conditions. This chapter will specifically discuss the regulation of Class IA PI 3-kinase enzymatic activity, focusing on regulatory interactions between the p85 and p110 subunits and the modulation of these interactions by physiological activators and oncogenic mutations. The complex web of signaling downstream from Class IA PI 3-kinases will be discussed in other chapters in this volume.
Figures
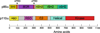
References
-
- Ahmed S, Lee J, Wen L-P, Zhao Z, Ho J, Best A, Kozma R, Lim L. Breakpoint Cluster Region gene product-related domain of n-Chimaerin. J Biol Chem. 1994;269:17642–17648. - PubMed
-
- Beeton CA, Das P, Waterfield MD, Shepherd PR. The SH3 and BH domains of the p85alpha adapter subunit play a critical role in regulating Class Ia phosphoinositide 3-kinase function. Mol Cell Biol Res Comm. 1999;1:153–157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

