The complete inventory of receptors encoded by the rat natural killer cell gene complex
- PMID: 20544345
- PMCID: PMC2910302
- DOI: 10.1007/s00251-010-0455-y
The complete inventory of receptors encoded by the rat natural killer cell gene complex
Abstract
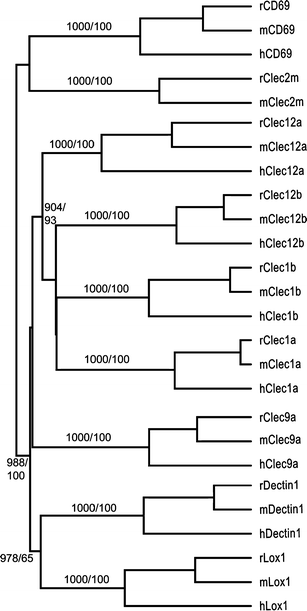
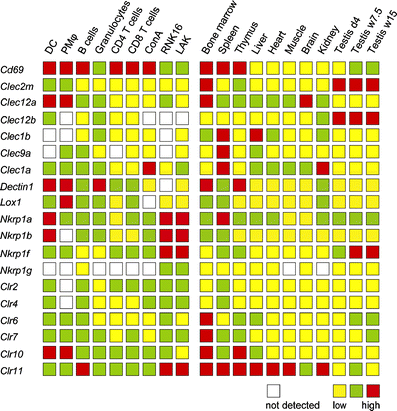
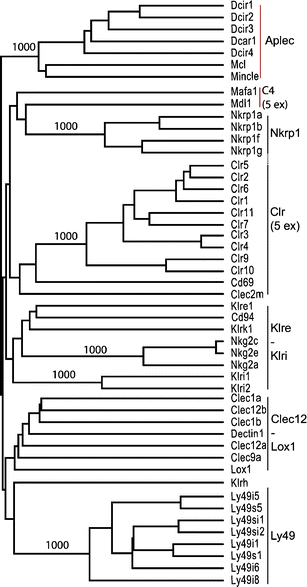
The natural killer cell gene complex (NKC) encodes receptors belonging to the C-type lectin superfamily expressed primarily by NK cells and other leukocytes. In the rat, the chromosomal region that starts with the Nkrp1a locus and ends with the Ly49i8 locus is predicted to contain 67 group V C-type lectin superfamily genes, making it one of the largest congregation of paralogous genes in vertebrates. Based on physical proximity and phylogenetic relationships between these genes, the rat NKC can be divided into four major parts. We have previously reported the cDNA cloning of the majority of the genes belonging to the centromeric Nkrp1/Clr cluster and the two telomeric groups, the Klre1-Klri2 and the Ly49 clusters. Here, we close the gap between the Nkrp1/Clr and the Klre1-Klri2 clusters by presenting the cDNA cloning and transcription patterns of eight genes spanning from Cd69 to Dectin1, including the novel Clec2m gene. The definition, organization, and evolution of the rat NKC are discussed.
Figures



Similar articles
-
Chicken CD69 and CD94/NKG2-like genes in a chromosomal region syntenic to mammalian natural killer gene complex.Immunogenetics. 2007 Jul;59(7):603-11. doi: 10.1007/s00251-007-0220-z. Epub 2007 May 16. Immunogenetics. 2007. PMID: 17505822
-
Cloning of Clr, a new family of lectin-like genes localized between mouse Nkrp1a and Cd69.Immunogenetics. 2001 Apr;53(3):209-14. doi: 10.1007/s002510100319. Immunogenetics. 2001. PMID: 11398965
-
Physiologic functions of activating natural killer (NK) complex-encoded receptors on NK cells.Immunol Rev. 2001 Jun;181:126-37. doi: 10.1034/j.1600-065x.2001.1810110.x. Immunol Rev. 2001. PMID: 11513134 Review.
-
Identification of natural killer cell receptor clusters in the platypus genome reveals an expansion of C-type lectin genes.Immunogenetics. 2009 Aug;61(8):565-79. doi: 10.1007/s00251-009-0386-7. Epub 2009 Jul 14. Immunogenetics. 2009. PMID: 19597809
-
The centromeric part of the human natural killer (NK) receptor complex: lectin-like receptor genes expressed in NK, dendritic and endothelial cells.Immunol Rev. 2001 Jun;181:5-19. doi: 10.1034/j.1600-065x.2001.1810101.x. Immunol Rev. 2001. PMID: 11513151 Review.
Cited by
-
Identification of a chicken CLEC-2 homologue, an activating C-type lectin expressed by thrombocytes.Immunogenetics. 2012 May;64(5):389-97. doi: 10.1007/s00251-011-0591-z. Epub 2011 Dec 29. Immunogenetics. 2012. PMID: 22205394
-
Dedicated immunosensing of the mouse intestinal epithelium facilitated by a pair of genetically coupled lectin-like receptors.Mucosal Immunol. 2015 Mar;8(2):232-42. doi: 10.1038/mi.2014.60. Epub 2014 Jul 2. Mucosal Immunol. 2015. PMID: 24985083
-
The c.503T>C Polymorphism in the Human KLRB1 Gene Alters Ligand Binding and Inhibitory Potential of CD161 Molecules.PLoS One. 2015 Aug 26;10(8):e0135682. doi: 10.1371/journal.pone.0135682. eCollection 2015. PLoS One. 2015. PMID: 26309225 Free PMC article.
-
C-Type Lectin-Like Receptors As Emerging Orchestrators of Sterile Inflammation Represent Potential Therapeutic Targets.Front Immunol. 2018 Feb 15;9:227. doi: 10.3389/fimmu.2018.00227. eCollection 2018. Front Immunol. 2018. PMID: 29497419 Free PMC article. Review.
-
Paired opposing leukocyte receptors recognizing rapidly evolving ligands are subject to homogenization of their ligand binding domains.Immunogenetics. 2011 Dec;63(12):809-20. doi: 10.1007/s00251-011-0553-5. Epub 2011 Jul 1. Immunogenetics. 2011. PMID: 21720914 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

