Functions of alternative replication protein A in initiation and elongation
- PMID: 20545304
- PMCID: PMC2912413
- DOI: 10.1021/bi100380n
Functions of alternative replication protein A in initiation and elongation
Abstract
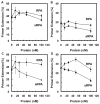
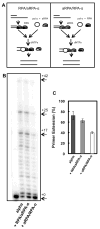
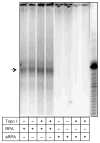
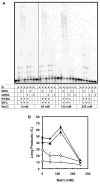
Replication protein A (RPA) is a single-stranded DNA-binding complex that is essential for DNA replication, repair, and recombination in eukaryotic cells. In addition to this canonical complex, we have recently characterized an alternative replication protein A complex (aRPA) that is unique to primates. aRPA is composed of three subunits: RPA1 and RPA3, also present in canonical RPA, and a primate-specific subunit RPA4, homologous to canonical RPA2. aRPA has biochemical properties similar to those of the canonical RPA complex but does not support DNA replication. We describe studies that aimed to identify what properties of aRPA prevent it from functioning in DNA replication. We show aRPA has weakened interaction with DNA polymerase alpha (pol alpha) and that aRPA is not able to efficiently stimulate DNA synthesis by pol alpha on aRPA-coated DNA. Additionally, we show that aRPA is unable to support de novo priming by pol alpha. Because pol alpha activity is essential for both initiation and Okazaki strand synthesis, we conclude that the inability of aRPA to support pol alpha loading causes aRPA to be defective in DNA replication. We also show that aRPA stimulates synthesis by DNA polymerase alpha in the presence of PCNA and RFC. This indicates that aRPA can support extension of DNA strands by DNA polymerase partial differential. This finding along with the previous observation that aRPA supports early steps of nucleotide excision repair and recombination indicates that aRPA can support DNA repair synthesis that requires polymerase delta, PCNA, and RFC and support a role for aRPA in DNA repair.
Figures




Similar articles
-
An alternative form of replication protein a expressed in normal human tissues supports DNA repair.J Biol Chem. 2010 Feb 12;285(7):4788-97. doi: 10.1074/jbc.M109.079418. Epub 2009 Dec 7. J Biol Chem. 2010. PMID: 19996105 Free PMC article.
-
An alternative form of replication protein a prevents viral replication in vitro.J Biol Chem. 2009 Feb 20;284(8):5324-31. doi: 10.1074/jbc.M808963200. Epub 2008 Dec 29. J Biol Chem. 2009. PMID: 19116208 Free PMC article.
-
Replication protein A, the laxative that keeps DNA regular: The importance of RPA phosphorylation in maintaining genome stability.Semin Cell Dev Biol. 2019 Feb;86:112-120. doi: 10.1016/j.semcdb.2018.04.005. Epub 2018 Nov 13. Semin Cell Dev Biol. 2019. PMID: 29665433 Review.
-
Effect of 8-oxoguanine and abasic site DNA lesions on in vitro elongation by human DNA polymerase in the presence of replication protein A and proliferating-cell nuclear antigen.Biochem J. 2010 Aug 1;429(3):573-82. doi: 10.1042/BJ20100405. Biochem J. 2010. PMID: 20528769
-
Eukaryotic single-stranded DNA binding proteins: central factors in genome stability.Subcell Biochem. 2010;50:143-63. doi: 10.1007/978-90-481-3471-7_8. Subcell Biochem. 2010. PMID: 20012581 Review.
Cited by
-
The roles of telomerase in the generation of polyploidy during neoplastic cell growth.Neoplasia. 2013 Feb;15(2):156-68. doi: 10.1593/neo.121398. Neoplasia. 2013. PMID: 23441130 Free PMC article.
-
Genome Stability Maintenance in Naked Mole-Rat.Acta Naturae. 2017 Oct-Dec;9(4):31-41. Acta Naturae. 2017. PMID: 29340215 Free PMC article.
-
A slipped-CAG DNA-binding small molecule induces trinucleotide-repeat contractions in vivo.Nat Genet. 2020 Feb;52(2):146-159. doi: 10.1038/s41588-019-0575-8. Epub 2020 Feb 14. Nat Genet. 2020. PMID: 32060489 Free PMC article.
-
Shared Subunits of Tetrahymena Telomerase Holoenzyme and Replication Protein A Have Different Functions in Different Cellular Complexes.J Biol Chem. 2017 Jan 6;292(1):217-228. doi: 10.1074/jbc.M116.763664. Epub 2016 Nov 28. J Biol Chem. 2017. PMID: 27895115 Free PMC article.
-
Antagonistic roles of canonical and Alternative-RPA in disease-associated tandem CAG repeat instability.Cell. 2023 Oct 26;186(22):4898-4919.e25. doi: 10.1016/j.cell.2023.09.008. Epub 2023 Oct 11. Cell. 2023. PMID: 37827155 Free PMC article.
References
-
- Wold MS. Replication Protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. - PubMed
-
- Iftode C, Daniely Y, Borowiec JA. Replication Protein A (RPA): The eukaryotic SSB. CRC Critical Reviews in Biochemistry. 1999;34:141–180. - PubMed
-
- Rider SD, Jr, Cai X, Sullivan WJ, Jr, Smith AT, Radke J, White MGZ. The protozoan parasite Cryptosporidium parvum possesses two functionally and evolutionarily divergent replication protein A large subunits. Journal of Bological Chemistry. 2005;280:31460–31469. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

