Intracellular localization of membrane-bound ATPases in the compartmentalized anammox bacterium 'Candidatus Kuenenia stuttgartiensis'
- PMID: 20545867
- PMCID: PMC2936114
- DOI: 10.1111/j.1365-2958.2010.07242.x
Intracellular localization of membrane-bound ATPases in the compartmentalized anammox bacterium 'Candidatus Kuenenia stuttgartiensis'
Abstract
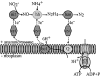
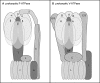
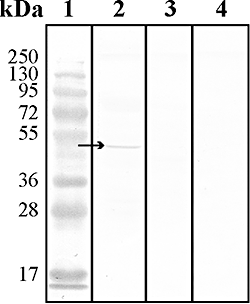
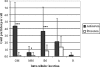
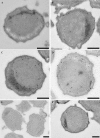
Anaerobic ammonium-oxidizing (anammox) bacteria are divided into three compartments by bilayer membranes (from out- to inside): paryphoplasm, riboplasm and anammoxosome. It is proposed that the anammox reaction is performed by proteins located in the anammoxosome and on its membrane giving rise to a proton-motive-force and subsequent ATP synthesis by membrane-bound ATPases. To test this hypothesis, we investigated the location of membrane-bound ATPases in the anammox bacterium 'Candidatus Kuenenia stuttgartiensis'. Four ATPase gene clusters were identified in the K. stuttgartiensis genome: one typical F-ATPase, two atypical F-ATPases and a prokaryotic V-ATPase. K. stuttgartiensis transcriptomic and proteomic analysis and immunoblotting using antisera directed at catalytic subunits of the ATPase gene clusters indicated that only the typical F-ATPase gene cluster most likely encoded a functional ATPase under these cultivation conditions. Immunogold localization showed that the typical F-ATPase was predominantly located on both the outermost and anammoxosome membrane and to a lesser extent on the middle membrane. This is consistent with the anammox physiology model, and confirms the status of the outermost cell membrane as cytoplasmic membrane. The occurrence of ATPase in the anammoxosome membrane suggests that anammox bacteria have evolved a prokaryotic organelle; a membrane-bounded compartment with a specific cellular function: energy metabolism.
Figures
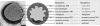

References
-
- Arrigo KR. Marine microorganisms and global nutrient cycles. Nature. 2005;437:349–355. - PubMed
-
- Boyer PD. The binding change mechanism for ATP synthase: some probabilities and possibilities. Biochim Biophys Acta. 1993;1140:215–250. - PubMed
-
- Boyer PD. The ATP synthase – a splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. - PubMed
-
- Brandes JA, Devol AH, Deutsch C. New developments in the marine nitrogen cycle. Chem Rev. 2007;107:577–589. - PubMed
-
- Cirpus IEY, de Been M, Op den Camp HJM, Strous M, Le Paslier D, Kuenen JG, Jetten MSM. A new soluble 10 kDa monoheme cytochrome c-552 from the anammox bacterium Candidatus‘Kuenenia stuttgartiensis’. FEMS Microbiol Lett. 2005;252:273–278. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

